6M1D
 
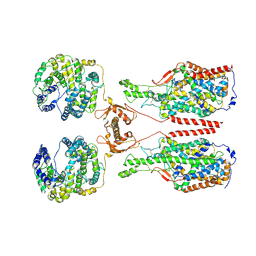 | | ACE2-B0AT1 complex, open conformation | | Descriptor: | Angiotensin-converting enzyme 2, Sodium-dependent neutral amino acid transporter B(0)AT1 | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
8D36
 
 | |
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8GRS
 
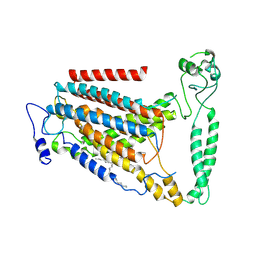 | | human TMEM63A | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CSC1-like protein 1 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
8GRN
 
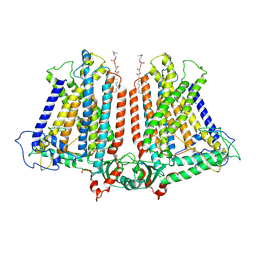 | | AtOSCA1.1 extended state | | Descriptor: | Protein OSCA1, [1-MYRISTOYL-GLYCEROL-3-YL]PHOSPHONYLCHOLINE | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
8GSO
 
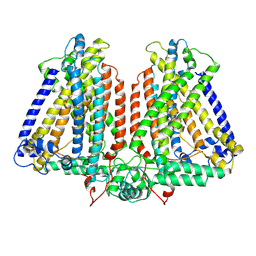 | | AtOSCA3.1 channel extended state | | Descriptor: | CSC1-like protein ERD4 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
8GRO
 
 | | AtOSCA3.1 contracted state | | Descriptor: | CSC1-like protein ERD4 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
5JRS
 
 | |
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
7P13
 
 | | 2.29 A Mycobacterium tuberculosis EspB. | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Gijsbers, A, Zhang, Y, Vinciauskaite, V, Siroy, A, Gao, Y, Tria, G, Mathew, A, Sanchez-Puig, N, Lopez-Iglesias, C, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Priming mycobacterial ESX-secreted protein B to form a channel-like structure.
Curr Res Struct Biol, 3, 2021
|
|
5M4Y
 
 | |
1G72
 
 | | CATALYTIC MECHANISM OF QUINOPROTEIN METHANOL DEHYDROGENASE: A THEORETICAL AND X-RAY CRYSTALLOGRAPHIC INVESTIGATION | | Descriptor: | CALCIUM ION, METHANOL DEHYDROGENASE HEAVY SUBUNIT, METHANOL DEHYDROGENASE LIGHT SUBUNIT, ... | | Authors: | Zheng, Y, Xia, Z, Chen, Z, Bruice, T.C, Mathews, F.S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of quinoprotein methanol dehydrogenase: A theoretical and x-ray crystallographic investigation.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8BNS
 
 | |
8QNI
 
 | |
8QNG
 
 | |
8QNH
 
 | | Crystal structure of the E3 ubiquitin ligase Cbl-b with an allosteric inhibitor (WO2020264398 Ex23) | | Descriptor: | 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Discovery of a Novel Benzodiazepine Series of Cbl-b Inhibitors for the Enhancement of Antitumor Immunity.
Acs Med.Chem.Lett., 14, 2023
|
|
7UD7
 
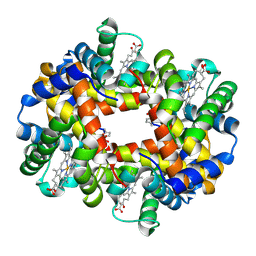 | | Crystal structure of deoxygenated hemoglobin in complex with 5HMF-NO at 1.8 Angstrom | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Donkor, A.K, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-18 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Antisickling Investigation of a Nitric Oxide-Releasing Prodrug of 5HMF for the Treatment of Sickle Cell Disease.
Biomolecules, 12, 2022
|
|
6TY9
 
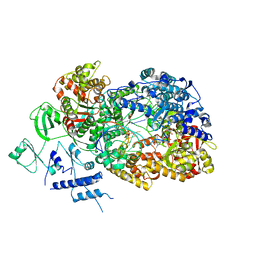 | | In situ structure of BmCPV RNA dependent RNA polymerase at initiation state | | Descriptor: | MAGNESIUM ION, Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*AP*AP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), RNA-dependent RNA Polymerase, ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ1
 
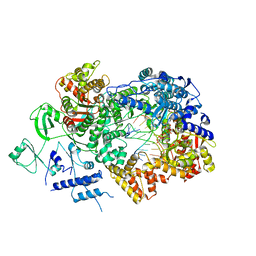 | | In situ structure of BmCPV RNA-dependent RNA polymerase at early-elongation state | | Descriptor: | Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*A)-3'), RNA-dependent RNA Polymerase, Template RNA (5'-R(P*AP*GP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TY8
 
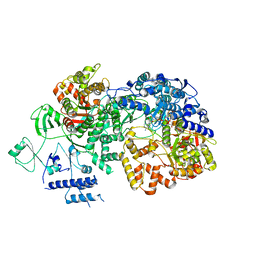 | | In situ structure of BmCPV RNA dependent RNA polymerase at quiescent state | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-dependent RNA Polymerase, Viral structural protein 4 | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7UD8
 
 | | Crystal structure of carbon monoxy Hemoglobin in complex with 5HMF at 1.8 Angstrom | | Descriptor: | (5-methylfuran-2-yl)methanol, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Donkor, A.K, Musayev, F.N, Safo, M.S. | | Deposit date: | 2022-03-18 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Antisickling Investigation of a Nitric Oxide-Releasing Prodrug of 5HMF for the Treatment of Sickle Cell Disease.
Biomolecules, 12, 2022
|
|
6TZ2
 
 | | In situ structure of BmCPV RNA-dependent RNA polymerase at elongation state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-template RNA (36-MER), ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
