5XXY
 
 | |
7X8M
 
 | | NMR Solution Structure of the 2:1 Berberine-KRAS-G4 Complex | | Descriptor: | BERBERINE, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8O
 
 | | NMR Solution Structure of the 2:1 Coptisine-KRAS-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8N
 
 | | NMR Solution Structure of the Wild-type Bulge-containing KRAS-G4 | | Descriptor: | DNA (24-mer) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
6UCE
 
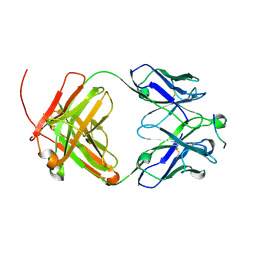 | |
7CMZ
 
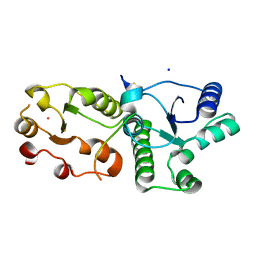 | | Crystal Structure of BRCT7/8 in Complex with the APS Motif of PHF8 | | Descriptor: | DNA topoisomerase 2-binding protein 1, Histone lysine demethylase PHF8, POTASSIUM ION, ... | | Authors: | Che, S.Y, Ma, S, Cao, C, Yao, Z, Shi, L, Yang, N. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability.
Sci Adv, 7, 2021
|
|
6UCF
 
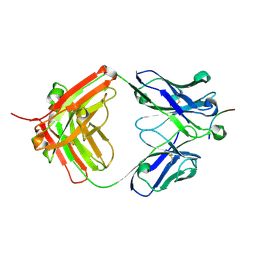 | |
5GZE
 
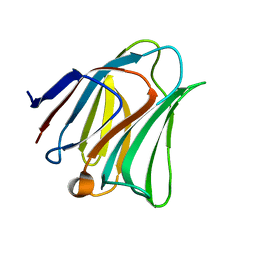 | | Galectin-8 N-terminal domain carbohydrate recognition domain | | Descriptor: | Galectin-8, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
5WVI
 
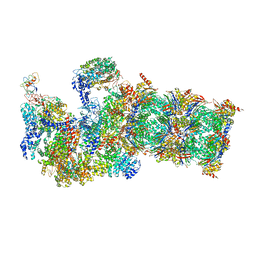 | | The resting state of yeast proteasome | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5EOA
 
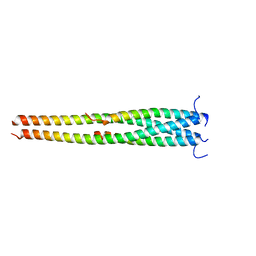 | | Crystal structure of OPTN E50K mutant and TBK1 complex | | Descriptor: | Optineurin, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
8KA8
 
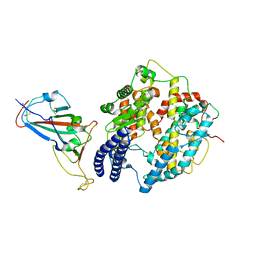 | | Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
5WVK
 
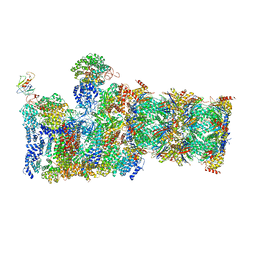 | | Yeast proteasome-ADP-AlFx | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
8KC2
 
 | | Cryo-EM structure of SARS-CoV-2 BA.3 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
5GZG
 
 | | Galectin-8 N-terminal domain carbohydrate recognition domain | | Descriptor: | Galectin-8, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
8UTE
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 27 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S)-6,6-difluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
5GZC
 
 | | Crystal structure of Galectin-8 N-CRD with part of linker | | Descriptor: | GLYCEROL, Galectin-8, SODIUM ION | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
7DGB
 
 | |
7DGH
 
 | |
7DGF
 
 | |
7DGG
 
 | |
7DGI
 
 | |
5GZD
 
 | | Galectin-8 N-terminal domain carbohydrate recognition domain | | Descriptor: | Galectin-8, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
8UPV
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPW
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 34 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6S)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
