2ZXD
 
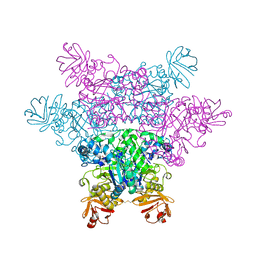 | | alpha-L-fucosidase complexed with inhibitor, iso-6FNJ | | Descriptor: | (2S,3R,4S,5R)-2-(1-methylethyl)piperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX7
 
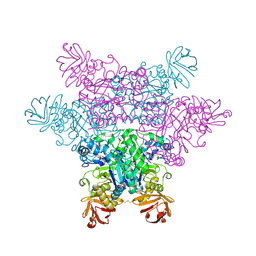 | | alpha-L-fucosidase complexed with inhibitor, F10-2C | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}-1H-indole-2-carboxamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX8
 
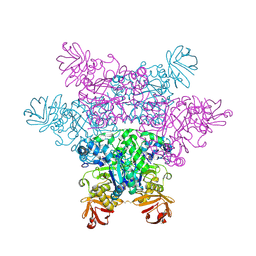 | | alpha-L-fucosidase complexed with inhibitor, F10-2C-O | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}-1-benzofuran-2-carboxamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
4DNB
 
 | | METHYLATION OF THE ECORI RECOGNITION SITE DOES NOT ALTER DNA CONFORMATION. THE CRYSTAL STRUCTURE OF D(CGCGAM6ATTCGCG) AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*(6MA)P*TP*TP*CP*GP*CP*G)-3') | | Authors: | Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Methylation of the EcoRI recognition site does not alter DNA conformation: the crystal structure of d(CGCGAm6ATTCGCG) at 2.0-A resolution.
J.Biol.Chem., 263, 1988
|
|
2D45
 
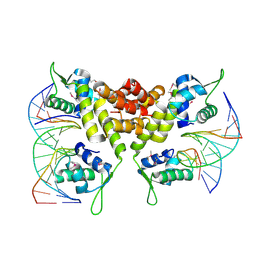 | | Crystal structure of the MecI-mecA repressor-operator complex | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*AP*TP*AP*TP*GP*TP*AP*GP*TP*A)-3', Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Ko, T.-P, Musayev, F.N, Zhao, Q, Wang, A.H.-J, Archer, G.L. | | Deposit date: | 2005-10-09 | | Release date: | 2005-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the MecI repressor from Staphylococcus aureus in complex with the cognate DNA operator of mec.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1UCS
 
 | | Type III Antifreeze Protein RD1 from an Antarctic Eel Pout | | Descriptor: | Antifreeze peptide RD1 | | Authors: | Ko, T.-P, Robinson, H, Gao, Y.-G, Cheng, C.-H.C, DeVries, A.L, Wang, A.H.-J. | | Deposit date: | 2003-04-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.62 Å) | | Cite: | The refined crystal structure of an eel pout type III antifreeze protein RD1 at 0.62-A resolution reveals structural microheterogeneity of protein and solvation.
Biophys.J., 84, 2003
|
|
2DND
 
 | | A BIFURCATED HYDROGEN-BONDED CONFORMATION IN THE D(A.T) BASE PAIRS OF THE DNA DODECAMER D(CGCAAATTTGCG) AND ITS COMPLEX WITH DISTAMYCIN | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Coll, M, Frederick, C.A, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin.
Proc.Natl.Acad.Sci.USA, 84, 1987
|
|
1D10
 
 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
2DES
 
 | | INTERACTIONS BETWEEN MORPHOLINYL ANTHRACYCLINES AND DNA: THE CRYSTAL STRUCTURE OF A MORPHOLINO DOXORUBICIN BOUND TO D(CGTACG) | | Descriptor: | 3'-DESAMINO-3'-(2-METHOXY-4-MORPHOLINYL)-DOXORUBICIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Cirilli, M, Bachechi, F, Ughetto, G, Colonna, F.P, Capobianco, M.L. | | Deposit date: | 1993-03-16 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions between morpholinyl anthracyclines and DNA. The crystal structure of a morpholino doxorubicin bound to d(CGTACG).
J.Mol.Biol., 230, 1993
|
|
381D
 
 | | BINDING OF THE MODIFIED DAUNORUBICIN WP401 ADJACENT TO A T-G BASE PAIR INDUCES THE REVERSE WATSON-CRICK CONFORMATION: CRYSTAL STRUCTURES OF THE WP401-TGGCCG AND WP401-CGG[BR5C]CG COMPLEXES | | Descriptor: | 2'-BROMO-4'-EPIDAUNORUBICIN, DNA (5'-D(*TP*GP*(G49)P*CP*CP*G)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*G)-3') | | Authors: | Dutta, R, Gao, Y.-G, Priebe, W, Wang, A.H.-J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the modified daunorubicin WP401 adjacent to a T-G base pair induces the reverse Watson-Crick conformation: crystal structures of the WP401-TGGCCG and WP401-CGG[br5C]CG complexes.
Nucleic Acids Res., 26, 1998
|
|
2OPN
 
 | | Human Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-527 | | Descriptor: | Farnesyl pyrophosphate synthetase (FPP synthetase) (FPS) (Farnesyl diphosphate synthetase) [Includes: Dimethylallyltranstransferase (EC 2.5.1.1); Geranyltranstransferase (EC 2.5.1.10)], MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Cao, R, Gao, Y.G, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2007-01-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bisphosphonates: Teaching Old Drugs with New Tricks
To be Published
|
|
1D53
 
 | | CRYSTAL STRUCTURE AT 1.5 ANGSTROMS RESOLUTION OF D(CGCICICG), AN OCTANUCLEOTIDE CONTAINING INOSINE, AND ITS COMPARISON WITH D(CGCG) AND D(CGCGCG) STRUCTURES | | Descriptor: | DNA (5'-D(*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*IP*CP*IP*CP*G)-3') | | Authors: | Kumar, V.D, Harrison, R.W, Andrews, L.C, Weber, I.T. | | Deposit date: | 1992-11-05 | | Release date: | 1993-04-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure at 1.5-A resolution of d(CGCICICG), an octanucleotide containing inosine, and its comparison with d(CGCG) and d(CGCGCG) structures.
Biochemistry, 31, 1992
|
|
3AMH
 
 | | crystal structure of cellulase 12A from Thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMP
 
 | | E134C-Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMQ
 
 | | E134C-Cellobiose co-crystal of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMM
 
 | | Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMN
 
 | | E134C-Cellobiose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
1D44
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1D12
 
 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), DOXORUBICIN, SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
1D14
 
 | | STRUCTURE OF 11-DEOXYDAUNOMYCIN BOUND TO DNA CONTAINING A PHOSPHOROTHIOATE | | Descriptor: | 6-DEOXYDAUNOMYCIN, DNA (5'-D(*CP*GP*TP*(AS)P*CP*G)-3') | | Authors: | Williams, L.D, Egli, M, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Quigley, G.J, Wang, A.H.-J, Rich, A, Frederick, C.A. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 11-deoxydaunomycin bound to DNA containing a phosphorothioate.
J.Mol.Biol., 215, 1990
|
|
1D86
 
 | | STRUCTURAL CONSEQUENCES OF A CARCINOGENIC ALKYLATION LESION ON DNA: EFFECT OF O6-ETHYL-GUANINE ON THE MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG)-NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1992-08-24 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural consequences of a carcinogenic alkylation lesion on DNA: effect of O6-ethylguanine on the molecular structure of the d(CGC[e6G]AATTCGCG)-netropsin complex.
Biochemistry, 31, 1992
|
|
1DNO
 
 | | A-DNA/RNA DODECAMER R(GCG)D(TATACGC) MG BINDING SITES | | Descriptor: | DNA/RNA (5'-R(*GP*CP*GP)-D(*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-16 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
1DCR
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-11-05 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1D9R
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', COBALT HEXAMMINE(III) | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-10-29 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1DNZ
 
 | | A-DNA DECAMER ACCGGCCGGT WITH MAGNESIUM BINDING SITES | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-17 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
