7ZQ9
 
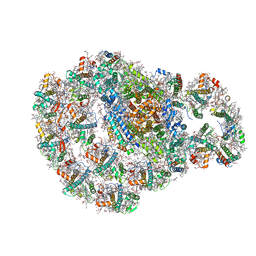 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZQE
 
 | |
7ZQC
 
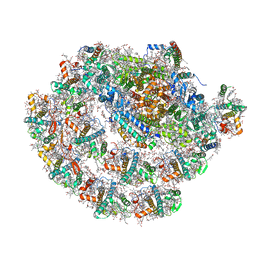 | | Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZQD
 
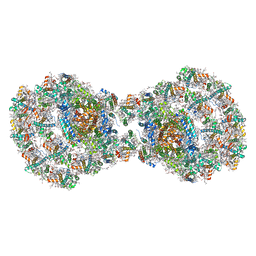 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7EA6
 
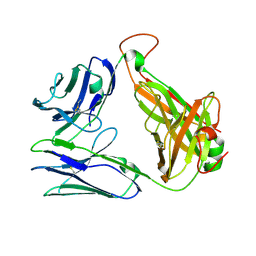 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
3GTU
 
 | |
8XQC
 
 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated PAF15 and hemimethylated DNA analog | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Kikuchi, A, Hayashi, G, Kori, S, Arita, K. | | Deposit date: | 2024-01-05 | | Release date: | 2025-01-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Activation of DNMT1 by Chemically Synthesized Dual monoubiquitinated PAF15 Protein
To Be Published
|
|
8YK4
 
 | | Structure of SARS-CoV-2 RBD and antibody NT-108 | | Descriptor: | Spike protein S1, antibody NT-108, single chain Fv fragment | | Authors: | Anraku, Y, Kita, S, Onodera, T, Sato, A, Tadokoro, T, Adachi, Y, Ito, S, Suzuki, T, Sasaki, J, Shiwa, N, Iwata, N, Nagata, N, Kazuki, Y, Oshimura, M, Sasaki, M, Orba, Y, Suzuki, T, Sawa, H, Hashiguchi, T, Fukuhara, H, Takahashi, Y, Maenaka, K. | | Deposit date: | 2024-03-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Potent Neutralization Activity of SARS-CoV-2 Antibody, NT-108
To Be Published
|
|
8YYM
 
 | | Cryo-EM structure of cylindrical fiber of MyD88 TIR | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Kasai, K, Imamura, K, Narita, A, Makino, F, Miyata, T, Kato, T, Namba, K, Onishi, H, Tochio, H. | | Deposit date: | 2024-04-04 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | From Monomers to Oligomers: Structural Mechanism of Receptor-Triggered MyD88 Assembly in Innate Immune Signaling
Biorxiv, 2024
|
|
8Y6X
 
 | | Crystal structure of ternary complex of human MR1, ligand #4, and MAIT-TCR A-F7 | | Descriptor: | 5-(2-oxidanylidenepropyl)-8-[(2~{S},3~{S},4~{R})-2,3,4,5-tetrakis(oxidanyl)pentyl]-1,7-dihydropteridine-2,4,6-trione, Beta-2-microglobulin, MAIT T cell receptor (A-F7) alpha chain, ... | | Authors: | Nagae, M, Inuki, S, Yamasaki, S. | | Deposit date: | 2024-02-03 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Development of Ribityllumazine Analogue as Mucosal-Associated Invariant T Cell Ligands.
J.Am.Chem.Soc., 146, 2024
|
|
7DKJ
 
 | |
8YKH
 
 | | Structure of SARS-CoV-2 spike RBD in complex with NT-108 scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-108 scFv, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Onodera, T, Tadokoro, T, Ito, S, Adachi, Y, Kotaki, R, Suzuki, T, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2024-03-05 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis for potent neutralization activity of SARS-CoV-2 antibody, NT-108
To Be Published
|
|
8YKG
 
 | | Structure of SARS-CoV-2 spike glycoprotein in complex with NT-108 scFv (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-108 scFv, ... | | Authors: | Anraku, Y, Kita, S, Onodera, T, Tadokoro, T, Ito, S, Adachi, Y, Kotaki, R, Suzuki, T, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2024-03-05 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for potent neutralization activity of SARS-CoV-2 antibody, NT-108
To Be Published
|
|
5YSD
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSF
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSE
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
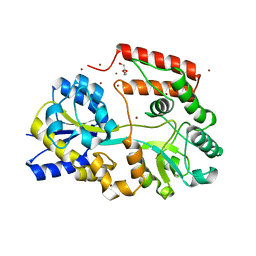 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
7CEP
 
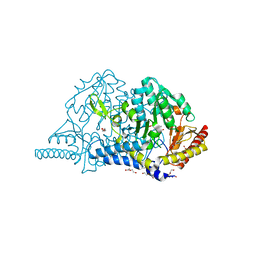 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Yasuhiro, T, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
6JZW
 
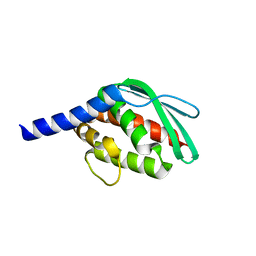 | |
6JZV
 
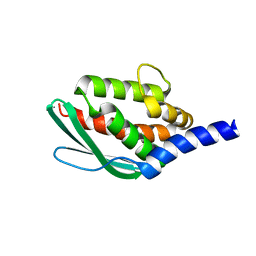 | | Crystal structure of SufU from Bacillus subtilis | | Descriptor: | ZINC ION, Zinc-dependent sulfurtransferase SufU | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cysteine-Persulfide Sulfane Sulfur-Ligated Zn Complex of Sulfur-Carrying SufU in the SufCDSUB System for Fe-S Cluster Biosynthesis.
Inorg.Chem., 2024
|
|
7DE4
 
 | | Hybrid cluster protein (HCP) from Escherichia coli | | Descriptor: | FE-S-O HYBRID CLUSTER, Hydroxylamine reductase, IRON/SULFUR CLUSTER | | Authors: | Fujishiro, T, Ooi, M, Takaoka, K, Takahashi, Y. | | Deposit date: | 2020-11-02 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Crystal structure of Escherichia coli class II hybrid cluster protein, HCP, reveals a [4Fe-4S] cluster at the N-terminal protrusion.
Febs J., 288, 2021
|
|
