8ENQ
 
 | | E. coli CsgA fibril (218-pixel box size) | | Descriptor: | Major curlin subunit | | Authors: | Bu, F, Liu, B. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into Escherichia coli CsgA amyloid fibril assembly.
Mbio, 15, 2024
|
|
8HD5
 
 | |
8THF
 
 | | SARS-CoV-2 BA.1 S-6P-no-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Bu, F, Li, F, Liu, B. | | Deposit date: | 2023-07-15 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Universal subunit vaccine protects against multiple SARS-CoV-2 variants and SARS-CoV.
Npj Vaccines, 9, 2024
|
|
1APL
 
 | | CRYSTAL STRUCTURE OF A MAT-ALPHA2 HOMEODOMAIN-OPERATOR COMPLEX SUGGESTS A GENERAL MODEL FOR HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*C P*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT-ALPHA2 HOMEODOMAIN) | | Authors: | Wolberger, C, Vershon, A.K, Liu, B, Johnson, A.D, Pabo, C.O. | | Deposit date: | 1993-10-04 | | Release date: | 1993-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 67, 1991
|
|
7XIN
 
 | | Crystal structure of DODC from Pseudomonas | | Descriptor: | DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Li, X, Zhou, Y.L, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Sun, D.Y, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of DODC from Pseudomonas
To Be Published
|
|
7XIO
 
 | | Crystal structure of TYR from Ralstonia | | Descriptor: | PHOSPHATE ION, Polyphenol oxidase | | Authors: | Sun, D.Y, Cui, P.P, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Zhang, J, Li, X, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of TYR from Ralstonia
To Be Published
|
|
9CO9
 
 | | Local refinement of JN.1 spike/Nanosota-9 complex | | Descriptor: | Nanosota-9, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
1YUK
 
 | | The crystal structure of the PSI/Hybrid domain/ I-EGF1 segment from the human integrin beta2 at 1.8 resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Integrin beta-2 A chain, Integrin beta-2 B chain | | Authors: | Shi, M, Sundramurthy, K, Liu, B, Tan, S.M, Law, S.K, Lescar, J. | | Deposit date: | 2005-02-14 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of the Plexin-Semaphorin-Integrin Domain/Hybrid Domain/I-EGF1 Segment from the Human Integrin {beta}2 Subunit at 1.8-A Resolution
J.Biol.Chem., 280, 2005
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU5
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU3
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU6
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
9CO8
 
 | | JN.1 spike/Nanosota-9 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-9, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9CO6
 
 | | BA.5 spike/Nanosota-9 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-9, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9CO7
 
 | | Local refinement of BA.5 spike/Nanosota-9 complex | | Descriptor: | Nanosota-9, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
1STS
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE FCHPQNT-NH2 DIMER | | Descriptor: | FCHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
1STR
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE AC-CHPQNT-NH2 DIMER | | Descriptor: | AC-CHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
7TGW
 
 | | Omicron spike at 3.0 A (open form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ye, G, Liu, B, Li, F. | | Deposit date: | 2022-01-09 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain.
Nat Commun, 13, 2022
|
|
7TGY
 
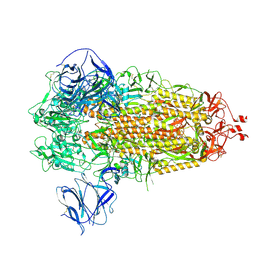 | | Prototypic SARS-CoV-2 G614 spike (closed form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ye, G, Liu, B, Li, F. | | Deposit date: | 2022-01-09 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain.
Nat Commun, 13, 2022
|
|
7TGX
 
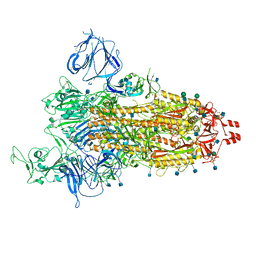 | | Prototypic SARS-CoV-2 G614 spike (open form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Ye, G, Liu, B, Li, F. | | Deposit date: | 2022-01-09 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain.
Nat Commun, 13, 2022
|
|
8K58
 
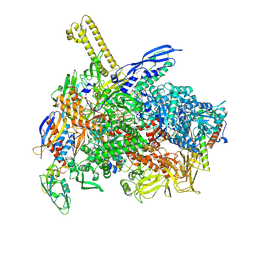 | | The cryo-EM map of close TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | TIEA inhibits Sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
8K5A
 
 | | The cryo-EM map of open TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
8K59
 
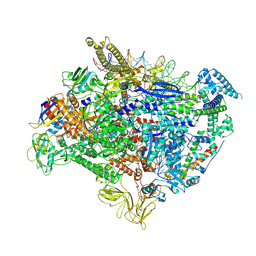 | | The cryo-EM map of TIC-TIEA complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (61-MER), DNA (63-MER), ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
