8Q4S
 
 | |
8QOB
 
 | |
7NPW
 
 | |
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS2
 
 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRX
 
 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS1
 
 | | Crystal structure of FAP R451K mutant in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7PU5
 
 | | Structure of SFPQ-NONO complex | | Descriptor: | MAGNESIUM ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Fribourg, S. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of SFPQ-NONO heterodimer.
Biochimie, 198, 2022
|
|
6TFQ
 
 | | Structure in P3212 form of the PBP/SBP MoaA in complex with mannopinic acid from A.tumefacien R10 | | Descriptor: | (2~{R})-2-[[(3~{R},4~{R},5~{S})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, ABC transporter substrate-binding protein, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
6TFX
 
 | | Structure in P21 form of the PBP/SBP MoaA in complex with mannopinic acid from A.tumefacien R10 | | Descriptor: | (2~{R})-2-[[(3~{R},4~{R},5~{S})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
6TG2
 
 | | Structure of the PBP/SBP MotA in complex with mannopinic acid from A.tumefacien R10 | | Descriptor: | (2~{R})-2-[[(3~{R},4~{R},5~{S})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Morera, S, Marty, L. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
6TFS
 
 | | Structure in P3212 form of the PBP/SBP MoaA in complex with glucopinic acid from A.tumefacien R10 | | Descriptor: | (2~{S})-2-[[(3~{S},4~{R},5~{R})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, ABC transporter substrate-binding protein, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
5OW4
 
 | |
7ZT6
 
 | |
7ZVT
 
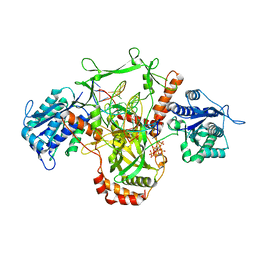 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
6R3Z
 
 | |
2X6N
 
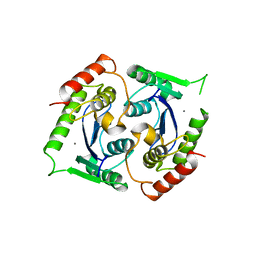 | | Human foamy virus integrase - catalytic core. Manganese-bound structure. | | Descriptor: | INTEGRASE, MANGANESE (II) ION | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-18 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2X6S
 
 | | Human foamy virus integrase - catalytic core. Magnesium-bound structure. | | Descriptor: | INTEGRASE, MAGNESIUM ION | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-19 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7Q5A
 
 | | Lanreotide nanotube | | Descriptor: | Lanreotide | | Authors: | Pieri, L, Wang, F, Arteni, A.A, Bressanelli, S, Egelman, E.H, Paternostre, M. | | Deposit date: | 2021-11-03 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Atomic structure of Lanreotide nanotubes revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6RU4
 
 | |
5M2W
 
 | | Structure of nanobody nb18 raised against TssK from E. coli T6SS | | Descriptor: | Llama nanobody nb8 against TssK from T6SS, SULFATE ION | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Desmyter, A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-28 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
6R5S
 
 | |
6R6K
 
 | | Structure of a FpvC mutant from pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique ferrous iron binding mode is associated with large conformational changes for the transport protein FpvC of Pseudomonas aeruginosa.
Febs J., 287, 2020
|
|
6R44
 
 | |
