5REK
 
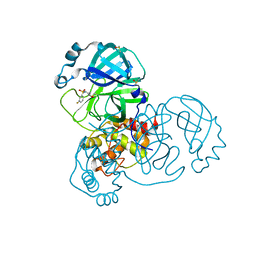 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102327 | | Descriptor: | 1-{4-[(3-fluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RF0
 
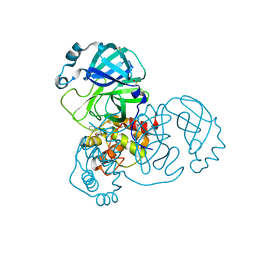 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with POB0073 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, [1-(pyridin-2-yl)cyclopentyl]methanol | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5ZVB
 
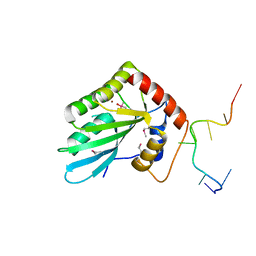 | | APOBEC3F Chimeric Catalytic Domain in Complex with DNA(dT9) | | Descriptor: | APEBEC3F/ssDNA-T9, CACODYLATE ION, DNA (5'-D(*AP*TP*TP*TP*TP*CP*AP*AP*T)-3'), ... | | Authors: | Cheng, C, Zhang, T.L, Wang, C.X, Lan, W.X, Ding, J.P, Cao, C.Y. | | Deposit date: | 2018-05-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Human APOBEC3F Chimeric Catalytic Domain in Complex with DNA
Chin.J.Chem., 36, 2018
|
|
5RFI
 
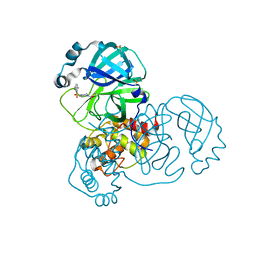 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102353 | | Descriptor: | 1-{4-[(2,5-dimethylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFW
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102243 | | Descriptor: | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
9FCT
 
 | |
5REA
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z31432226 | | Descriptor: | (azepan-1-yl)(2H-1,3-benzodioxol-5-yl)methanone, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RES
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102281 | | Descriptor: | 1-{4-[(2-fluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RF8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z271004858 | | Descriptor: | 3C-like proteinase, 4-amino-N-(pyridin-2-yl)benzenesulfonamide, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102179 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[3-(2-oxopyrrolidin-1-yl)phenyl]acetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4WF2
 
 | | Structure of E. coli BirA G142A bound to biotinol-5'-AMP | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Bifunctional ligase/repressor BirA | | Authors: | Eginton, C, Beckett, D, Wade, H. | | Deposit date: | 2014-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Allosteric Coupling via Distant Disorder-to-Order Transitions.
J.Mol.Biol., 427, 2015
|
|
4WFT
 
 | | Crystal structure of tRNA-dihydrouridine(20) synthase dsRBD domain | | Descriptor: | tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
4WHN
 
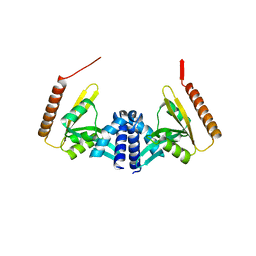 | | Structure of toxin-activating acyltransferase (TAAT) | | Descriptor: | ApxC, CITRIC ACID | | Authors: | Crow, A, Greene, N.P, Hughes, C, Koronakis, V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a bacterial toxin-activating acyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
9FVD
 
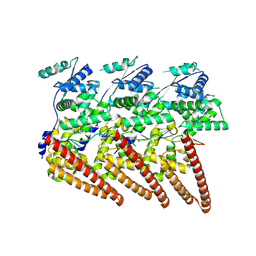 | | Cryo-EM structure of single-layered nucleoprotein-RNA helical assembly from Marburg virus, trimeric repeat unit | | Descriptor: | Nucleoprotein, RNA (5'-R(P*AP*GP*AP*CP*AP*CP*AP*CP*AP*AP*AP*AP*AP*CP*AP*AP*GP*A)-3') | | Authors: | Zinzula, L, Beck, F, Camasta, M, Bohn, S, Liu, C, Morado, D, Bracher, A, Plitzko, J.M, Baumeister, W. | | Deposit date: | 2024-06-26 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of single-layered nucleoprotein-RNA complex from Marburg virus.
Nat Commun, 15, 2024
|
|
9FYD
 
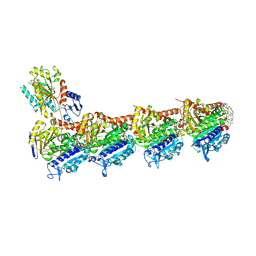 | | tubulin - cryptophycin-uD[Dab] complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dessin, C, Schachtsiek, T, Voss, J, Abel, A.-C, Neumann, B, Stammler, H.-G, Prota, A.E, Sewald, N. | | Deposit date: | 2024-07-03 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Highly Cytotoxic Cryptophycin Derivatives with Modification in Unit D for Conjugation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7OA3
 
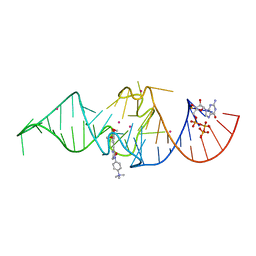 | | Crystal structure of Chili RNA aptamer in complex with DMHBO+ (Iridium hexammine co-crystallized form) | | Descriptor: | Chili RNA Aptamer, DMHBO+, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
5REJ
 
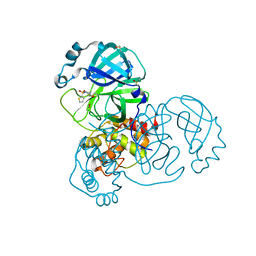 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102241 | | Descriptor: | 1-{4-[(thiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
7OAW
 
 | | Crystal structure of the Chili RNA aptamer in complex with DMHBI+ | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBI+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OAX
 
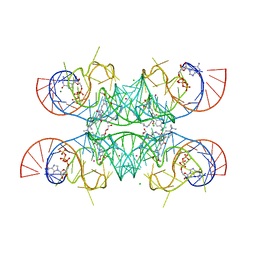 | | Crystal structure of the Chili RNA aptamer in complex with DMHBO+ | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBO+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
5REZ
 
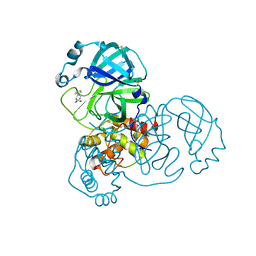 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with POB0129 | | Descriptor: | (1R,2S)-2-(thiophen-3-yl)cyclopentane-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
7OAV
 
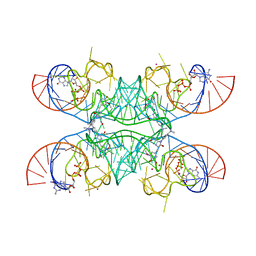 | | Crystal structure of Chili RNA aptamer in complex with DMHBO+ (Iridium III hexammine soaking crystal form) | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBO+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
5RFE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z509756472 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(4-cyanophenyl)methyl]morpholine-4-carboxamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFT
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102432 | | Descriptor: | 1-[(4S)-4-phenyl-3,4-dihydroisoquinolin-2(1H)-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4WR5
 
 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4WSE
 
 | |
