9JRM
 
 | | outward-open hSLC19A1 + 2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Soluble cytochrome b562,Reduced folate transporter,Soluble cytochrome b562,Reduced folate transporter,fusion protein | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
9JRL
 
 | | outward-open hSLC19A1 + PT523 | | Descriptor: | N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, Soluble cytochrome b562,Reduced folate transporter,fusion protein | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
9JRN
 
 | | outward-open hSLC19A1 + TPP | | Descriptor: | Soluble cytochrome b562,Reduced folate transporter,fusion protein, THIAMINE DIPHOSPHATE | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
9KNI
 
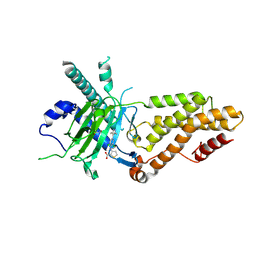 | | Structural complex of FTO bound with 12j | | Descriptor: | 2-OXOGLUTARIC ACID, 3-[[2,6-bis(chloranyl)-4-(3,5-dimethyl-1H-pyrazol-4-yl)phenyl]amino]thiophene-2-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.G, Gan, J.H. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Development of 3-arylaminothiophenic-2-carboxylic acid derivatives as new FTO inhibitors showing potent antileukemia activities.
Eur.J.Med.Chem., 289, 2025
|
|
5KL4
 
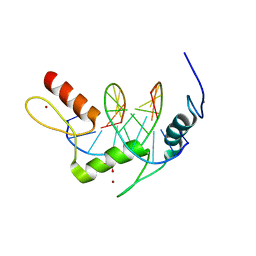 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with formylated DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KKQ
 
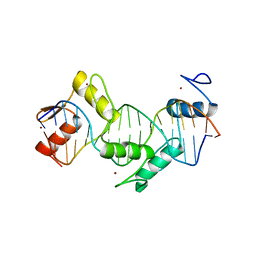 | | Homo sapiens CCCTC-binding factor (CTCF) ZnF3-7 and DNA complex structure | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5KL5
 
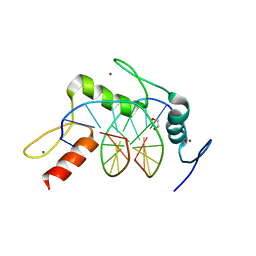 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with carboxylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KL2
 
 | | Wilms Tumor Protein (WT1) ZnF2-4 in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
1JRU
 
 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 (ENERGY MINIMISED AVERAGE) | | Descriptor: | p47 protein | | Authors: | Yuan, X.M, Shaw, A, Zhang, X.D, Kondo, H, Lally, J, Freemont, P.S, Matthews, S.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
6WE8
 
 | | YTH domain of human YTHDC1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
5GN8
 
 | | Structure of a 48-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | CALCIUM ION, Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Zhao, G, Mikami, B. | | Deposit date: | 2016-07-19 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | "Silent" Amino Acid Residues at Key Subunit Interfaces Regulate the Geometry of Protein Nanocages
ACS Nano, 10, 2016
|
|
4N0N
 
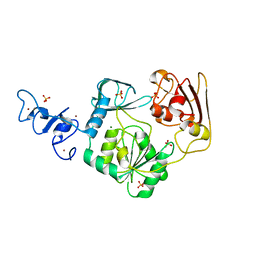 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | Descriptor: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
5XGH
 
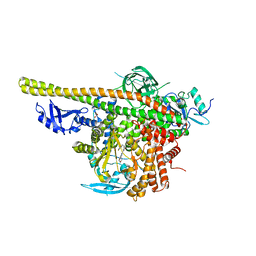 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
2L83
 
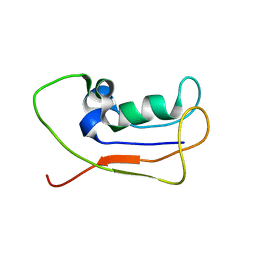 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
5G5J
 
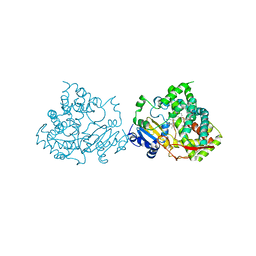 | | Crystal structure of human CYP3A4 bound to metformin | | Descriptor: | CYTOCHROME P450 3A4, Metformin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2016-05-25 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme Binding Biguanides Target Cytochrome P450-Dependent Cancer Cell Mitochondria.
Cell Chem Biol, 24, 2017
|
|
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Chen, Y.H, Wang, X.C, Zhang, C.R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7XPQ
 
 | |
7XPP
 
 | |
7L5M
 
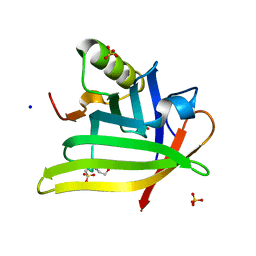 | | Crystal Structure of the DiB-RM-split Protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lipocalin family protein, SODIUM ION, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
7L5L
 
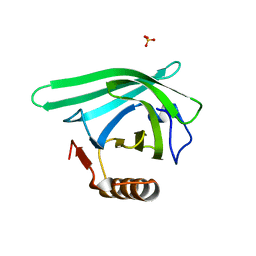 | |
7L5K
 
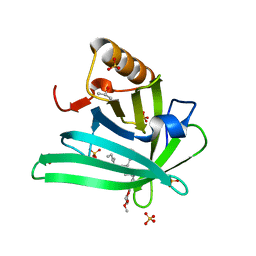 | | Crystal structure of the DiB-RM protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ISOPROPYL ALCOHOL, Lipocalin family protein, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
5KML
 
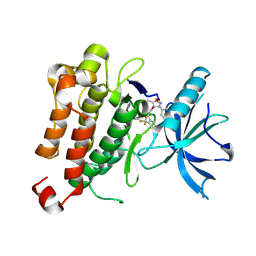 | |
5KMN
 
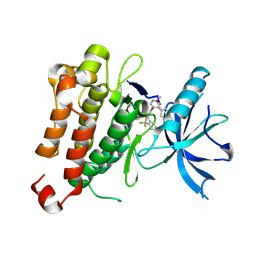 | |
5KMK
 
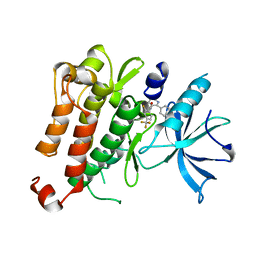 | |
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
