4X6N
 
 | |
6TUL
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021177 | | Descriptor: | D-MALATE, Fab C0021177 heavy chain (IgG1), Fab C0021177 light chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
3SBD
 
 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
6SRX
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
7B14
 
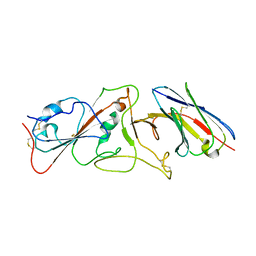 | | Nanobody E bound to Spike-RBD in a localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-04-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B18
 
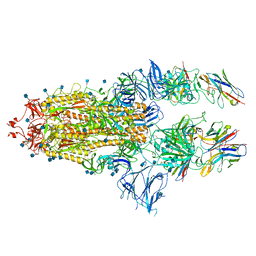 | | SARS-CoV-spike bound to two neutralising nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
5DNI
 
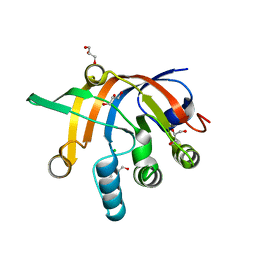 | | Crystal structure of Methanocaldococcus jannaschii Fumarate hydratase beta subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jayaraman, V, Kunala, J, Balaram, H. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Revisiting the Burden Borne by Fumarase: Enzymatic Hydration of an Olefin.
Biochemistry, 62, 2023
|
|
2FH7
 
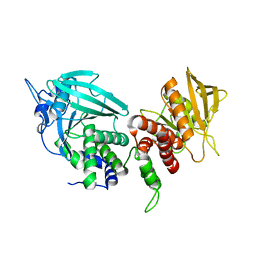 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1YDS
 
 | | Structure of CAMP-dependent protein kinase, alpha-catalytic subunit in complex with H8 protein kinase inhibitor [N-(2-methylamino)ethyl]-5-isoquinolinesulfonamide | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(METHYLAMINO)ETHYL]-5-ISOQUINOLINESULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDR
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H7 PROTEIN KINASE INHIBITOR 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE | | Descriptor: | 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE, C-AMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDT
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H89 PROTEIN KINASE INHIBITOR N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
1QXW
 
 | | Crystal structure of Staphyloccocus aureus in complex with an aminoketone inhibitor 54135. | | Descriptor: | (3S)-3-AMINO-1-(CYCLOPROPYLAMINO)HEPTANE-2,2-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
2ABD
 
 | |
3PIS
 
 | |
3QTL
 
 | |
6XDB
 
 | | Crystal structure of IRE1a in complex with G-6904 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-N-(6-chloro-2-fluoro-3-{[(2-fluorophenyl)sulfonyl]amino}phenyl)-6-(1,3-dimethyl-1H-pyrazol-4-yl)quinazoline-8-carboxamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.A, Weiru, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
6XDD
 
 | | Crystal structure of IRE1 in complex with G-3053 | | Descriptor: | 4-[(trans-4-aminocyclohexyl)amino]-N-(6-chloro-3-{[(2,5-difluorophenyl)sulfonyl]amino}-2-fluorophenyl)thieno[3,2-d]pyrimidine-7-carboxamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Ackerly-Wallweber, H, Wang, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
1WUT
 
 | | Acyl Ureas as Human Liver Glycogen Phosphorylase Inhibitors for the Treatment of Type 2 Diabetes | | Descriptor: | 7-[2,6-DICHLORO-4-({[(2-CHLOROBENZOYL)AMINO]CARBONYL}AMINO)PHENOXY]HEPTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.-J, Herling, A.W, Oikonomakos, N.G, Kosmopoulou, M.N, Schmoll, D, Sarubbi, E, von Roedern, E, Schonafinger, K, Defossa, E. | | Deposit date: | 2004-12-08 | | Release date: | 2005-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
6LXY
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[(2R)-2-fluoranyl-3-methyl-3-oxidanyl-butyl]-6-[(6-fluoranylpyrazolo[1,5-a]pyrimidin-5-yl)amino]-4-(propan-2-ylamino)pyridine-3-carboxamide, SULFATE ION | | Authors: | Ghosh, K, Bose, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-11-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of Nicotinamides as Potent and Selective IRAK4 Inhibitors with Efficacy in a Murine Model of Psoriasis.
Acs Med.Chem.Lett., 11, 2020
|
|
5AO1
 
 | |
1RXD
 
 | | Crystal structure of human protein tyrosine phosphatase 4A1 | | Descriptor: | protein tyrosine phosphatase type IVA, member 1; Protein tyrosine phosphatase IVA1 | | Authors: | Sun, J.P, Fedorov, A.A, Almo, S.C, Zhang, Z.Y, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-18 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
5AO0
 
 | |
5AO3
 
 | |
5AO4
 
 | |
