4ZTO
 
 | |
5AGO
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-mercaptoacetimidamido)pentanoic acid | | Descriptor: | (S)-2-AMINO-5-(2-MERCAPTOACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
5AGP
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-mercaptoacetimidamido)pentanoic acid | | Descriptor: | (S)-2-AMINO-5-(2-MERCAPTOACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
6LRB
 
 | |
5AGK
 
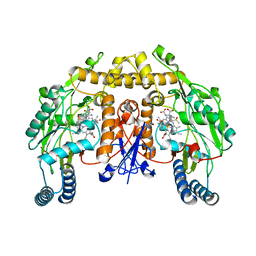 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (2S)-2-Amino-5-(2-(methylsulfinyl)acetimidamido) pentanoic acid | | Descriptor: | (2S)-2-AMINO-5-(2-(METHYLSULFINYL)ACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
3DFV
 
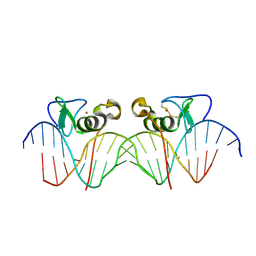 | | Adjacent GATA DNA binding | | Descriptor: | DNA (5'-D(*DAP*DAP*DGP*DCP*DAP*DGP*DAP*DTP*DAP*DAP*DGP*DTP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DG)-3'), DNA (5'-D(*DTP*DTP*DCP*DTP*DGP*DAP*DTP*DAP*DAP*DGP*DAP*DCP*DTP*DTP*DAP*DTP*DCP*DTP*DGP*DC)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Bates, D.L, Kim, G.K, Guo, L, Chen, L. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA.
J.Mol.Biol., 381, 2008
|
|
7K1B
 
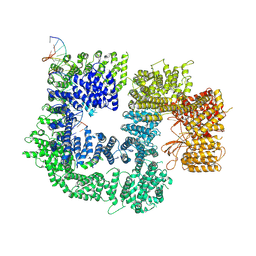 | | CryoEM structure of DNA-PK catalytic subunit complexed with DNA (Complex II) | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
5C8N
 
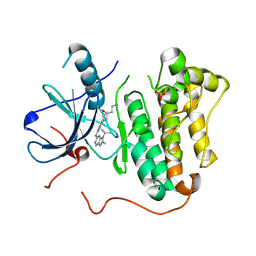 | | EGFR kinase domain mutant "TMLR" with compound 23 | | Descriptor: | Epidermal growth factor receptor, N-{2-[4-(2-aminoethyl)-4-methoxypiperidin-1-yl]pyrimidin-4-yl}-2-methyl-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-amine, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
7K11
 
 | |
7K0Y
 
 | | Cryo-EM structure of activated-form DNA-PK (complex VI) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-06 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K10
 
 | |
7K19
 
 | | CryoEM structure of DNA-PK catalytic subunit complexed with DNA (Complex I) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
5HCZ
 
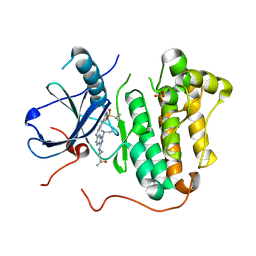 | | EGFR kinase domain mutant "TMLR" with 3-azetidinyl azaindazole compound 21 | | Descriptor: | 2-[1-[1-[(2~{S})-butan-2-yl]-6-[[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]amino]pyrazolo[4,3-c]pyridin-3-yl]azetidin-3-yl]propan-2-ol, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of a Noncovalent, Mutant-Selective Epidermal Growth Factor Receptor Inhibitor.
J.Med.Chem., 59, 2016
|
|
7K1J
 
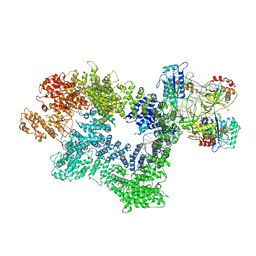 | | CryoEM structure of inactivated-form DNA-PK (Complex III) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
4ZTP
 
 | |
7K1N
 
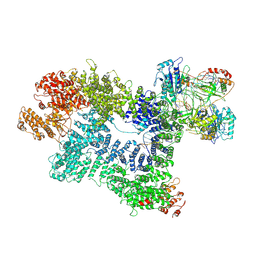 | | CryoEM structure of inactivated-form DNA-PK (Complex V) | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-08 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K17
 
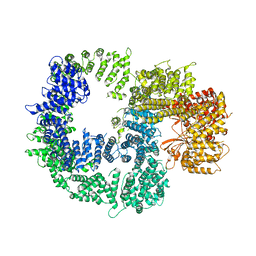 | |
5AGM
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-oxoacetimidamido)pentanoic acid | | Descriptor: | (S)-2-AMINO-5-(2-OXOACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
5CAN
 
 | | EGFR kinase domain mutant "TMLR" with compound 27 | | Descriptor: | (3R)-3-methyl-1-(4-{[2-methyl-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-yl]amino}pyrimidin-2-yl)pyrrolidine-3-carboxamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
5CAV
 
 | | EGFR kinase domain with compound 41a | | Descriptor: | (1R)-1-{6-({2-[(3R,4S)-3-fluoro-4-methoxypiperidin-1-yl]pyrimidin-4-yl}amino)-1-[(2S)-1,1,1-trifluoropropan-2-yl]-1H-imidazo[4,5-c]pyridin-2-yl}ethanol, Epidermal growth factor receptor | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
5AGN
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-hydroxyacetimidamido)pentanoic acid | | Descriptor: | (S)-2-AMINO-5-(2-HYDROXYACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
5AGL
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-(methylsulfonyl)acetimidamido)pentanoic acid | | Descriptor: | (S)-2-Amino-5-(2-(methylsulfonyl)acetimidamido)pentanoic acid, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
5C8M
 
 | | EGFR kinase domain mutant "TMLR" with compound 17 | | Descriptor: | 2-methyl-N-{2-[4-(methylsulfonyl)piperidin-1-yl]pyrimidin-4-yl}-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-amine, Epidermal growth factor receptor | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
5CAU
 
 | | EGFR kinase domain mutant "TMLR" with compound 41b | | Descriptor: | (1R)-1-{6-({2-[(3S,4R)-3-fluoro-4-methoxypiperidin-1-yl]pyrimidin-4-yl}amino)-1-[(2S)-1,1,1-trifluoropropan-2-yl]-1H-imidazo[4,5-c]pyridin-2-yl}ethanol, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
5JRE
 
 | | Crystal structure of NeC3PO in complex with ssDNA. | | Descriptor: | 9-METHYL-9H-PURIN-6-AMINE, ADENINE, NEQ131, ... | | Authors: | Gan, J, Zhang, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
