7UL0
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7WBO
 
 | | Crystal structure of Sarcoplasmic Calcium-Binding Protein from Scylla paramamosain | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Chen, Y, Jin, T, Liu, G. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure Analysis of Sarcoplasmic-Calcium-Binding Protein: An Allergen in Scylla paramamosain.
J.Agric.Food Chem., 71, 2023
|
|
7FJD
 
 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJE
 
 | | Cryo-EM structure of a membrane protein(LL) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJF
 
 | | Cryo-EM structure of a membrane protein(CS) | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7CVH
 
 | | Human Fructose-1,6-bisphosphatase 1 in complex with geranylgeranyl diphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structural basis for GGPP activation on the enzymatic activities FBP1
To Be Published
|
|
7D1G
 
 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase GAPDH from Clostridium beijerinckii | | Descriptor: | BETA-MERCAPTOETHANOL, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION | | Authors: | Chen, Y, Lan, J, Liu, W, Wang, L, Xu, Y. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase GAPDH from Clostridium beijerinckii
To Be Published
|
|
7D50
 
 | | SpuA mutant - H221N with glutamyl-thioester | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7X3K
 
 | | Cryo-EM structure of RAC in the State C2 RNC-RAC complex | | Descriptor: | Ribosome-associated complex subunit SSZ1, Zuotin | | Authors: | Chen, Y, Gao, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural remodeling of ribosome associated Hsp40-Hsp70 chaperones during co-translational folding.
Nat Commun, 13, 2022
|
|
7X34
 
 | |
7D54
 
 | | Crstal structure MsGATase with Gln | | Descriptor: | GLUTAMINE, Glutamine amidotransferase class-I | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D4R
 
 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D53
 
 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1G6M
 
 | |
1JE9
 
 | | NMR SOLUTION STRUCTURE OF NT2 | | Descriptor: | SHORT NEUROTOXIN II | | Authors: | Cheng, Y, Wang, W, Wang, J. | | Deposit date: | 2001-06-16 | | Release date: | 2001-07-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
1ZBH
 
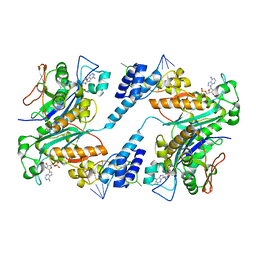 | | 3'-end specific recognition of histone mRNA stem-loop by 3'-exonuclease | | Descriptor: | 3'-5' exonuclease ERI1, 5'-R(*CP*CP*GP*GP*CP*UP*CP*UP*UP*UP*UP*CP*AP*GP*AP*GP*CP*CP*GP*G)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Cheng, Y, Patel, D.J. | | Deposit date: | 2005-04-08 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for 3'-end specific recognition of histone mRNA stem-loop by 3'-exonuclease, a human nuclease that also targets siRNA
To be Published
|
|
1ZBU
 
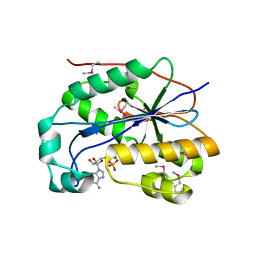 | | crystal structure of full-length 3'-exonuclease | | Descriptor: | 3'-5' exonuclease ERI1, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION | | Authors: | Cheng, Y, Patel, D.J. | | Deposit date: | 2005-04-08 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural basis for 3'-end specific recognition of histone mRNA stem-loop by 3'-exonuclease, a human nuclease that also targets siRNA
To be Published
|
|
1SUV
 
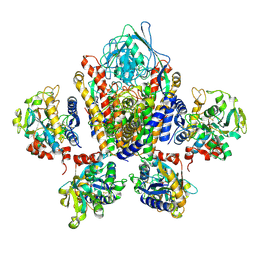 | | Structure of Human Transferrin Receptor-Transferrin Complex | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin, ... | | Authors: | Cheng, Y, Zak, O, Aisen, P, Harrison, S.C, Walz, T. | | Deposit date: | 2004-03-26 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the Human Transferrin Receptor-Transferrin Complex
Cell(Cambridge,Mass.), 116, 2004
|
|
1TJX
 
 | | Crystallographic Identification of Ca2+ Coordination Sites in Synaptotagmin I C2B Domain | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Cheng, Y, Sequeira, S.M, Malinina, L, Tereshko, V, Sollner, T.H, Patel, D.J. | | Deposit date: | 2004-06-07 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain.
Protein Sci., 13, 2004
|
|
1UOW
 
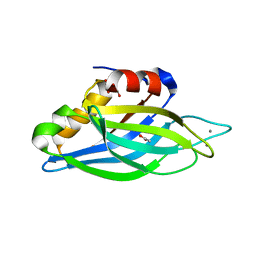 | | Calcium binding domain C2B | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Cheng, Y, Sequeira, S.M, Sollner, T.H, Patel, D.J. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystallographic Identification of Ca2+ and Sr2+ Coordination Sites in Synaptotagmin I C2B Domain
Protein Sci., 13, 2004
|
|
1UOV
 
 | | Calcium binding domain C2B | | Descriptor: | CALCIUM ION, GLYCEROL, SYNAPTOTAGMIN I | | Authors: | Cheng, Y, Sequeira, S.M, Sollner, T.H, Patel, D.J. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic Identification of Ca2+ and Sr2+ Coordination Sites in Synaptotagmin I C2B Domain
Protein Sci., 13, 2004
|
|
1TJM
 
 | | Crystallographic Identification of Sr2+ Coordination Site in Synaptotagmin I C2B Domain | | Descriptor: | GLYCEROL, STRONTIUM ION, Synaptotagmin I | | Authors: | Cheng, Y, Sequeira, S.M, Malinina, L, Tereshko, V, Sollner, T.H, Patel, D.J. | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain
Protein Sci., 13, 2004
|
|
1W0H
 
 | | Crystallographic structure of the nuclease domain of 3'hExo, a DEDDh family member, bound to rAMP | | Descriptor: | 3'-5' EXONUCLEASE ERI1, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION | | Authors: | Cheng, Y, Patel, D. | | Deposit date: | 2004-06-04 | | Release date: | 2004-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallographic Structure of the Nuclease Domain of 3'Hexo, a Deddh Family Member, Bound to Ramp
J.Mol.Biol., 343, 2004
|
|
3R0Q
 
 | |
3R8D
 
 | |
