3UB2
 
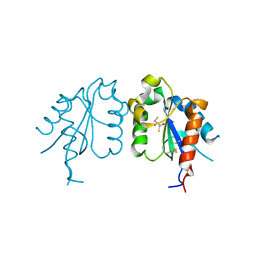 | | TIR domain of Mal/TIRAP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Shen, Y, Lin, Z. | | Deposit date: | 2011-10-23 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into TIR Domain Specificity of the Bridging Adaptor Mal in TLR4 Signaling
Plos One, 7, 2012
|
|
6EIL
 
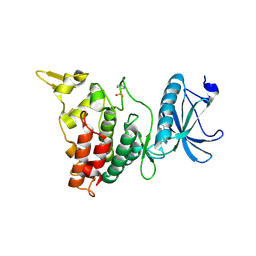 | | DYRK1A in complex with XMD8-49 | | Descriptor: | DYRK1A, [3-azanyl-6-(5-azanyl-2-methoxy-phenyl)pyrazin-2-yl]-pyridin-3-yl-methanone | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.465 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
2HD3
 
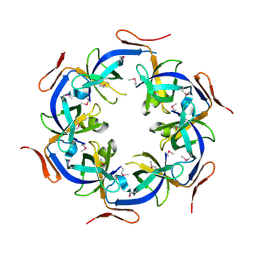 | | Crystal Structure of the Ethanolamine Utilization Protein EutN from Escherichia coli, NESG Target ER316 | | Descriptor: | Ethanolamine utilization protein eutN | | Authors: | Forouhar, F, Zhang, W, Jayaraman, S, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-19 | | Release date: | 2006-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1UEF
 
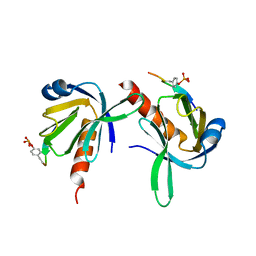 | | Crystal Structure of Dok1 PTB Domain Complex | | Descriptor: | 13-mer peptide from Proto-oncogene tyrosine-protein kinase receptor ret, Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yan, J, Rao, Z. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.Biol.Chem., 279, 2004
|
|
6ND2
 
 | | Staphylococcus aureus Dihydrofolate Reductase complexed with NADPH and 6-ETHYL-5-[(3R)-3-[2-METHOXY-5-(PYRIDIN-4-YL)PHENYL]BUT-1-YN-1-YL]PYRIMIDINE-2,4-DIAMINE (UCP1063) | | Descriptor: | 6-ethyl-5-{(3S)-3-[2-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Reeve, S.M, Wright, D.L. | | Deposit date: | 2018-12-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Structure-Guided In Vitro to In Vivo Pharmacokinetic Optimization of Propargyl-Linked Antifolates.
Drug Metab.Dispos., 47, 2019
|
|
1SQS
 
 | | X-Ray Crystal Structure Protein SP1951 of Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR27. | | Descriptor: | L(+)-TARTARIC ACID, conserved hypothetical protein | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM, 8, 2007
|
|
1GIU
 
 | | A TRICHOSANTHIN(TCS) MUTANT(E85R) COMPLEX STRUCTURE WITH ADENINE | | Descriptor: | ADENINE, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Guo, Q, Liu, Y, Dong, Y, Rao, Z. | | Deposit date: | 2001-03-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate binding and catalysis in trichosanthin occur in different sites as revealed by the complex structures of several E85 mutants.
Protein Eng., 16, 2003
|
|
1RTY
 
 | | Crystal Structure of Bacillus subtilis YvqK, a putative ATP-binding Cobalamin Adenosyltransferase, The North East Structural Genomics Target SR128 | | Descriptor: | PHOSPHATE ION, yvqk protein | | Authors: | Forouhar, F, Lee, I, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1TM0
 
 | | Crystal Structure of the putative proline racemase from Brucella melitensis, Northeast Structural Genomics Target LR31 | | Descriptor: | PROLINE RACEMASE | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Ho, C.K, Ma, L.-C, Cooper, B, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-10 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4HOG
 
 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 5-[3-(2-methoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine (UCP111H) | | Descriptor: | 5-[3-(2-methoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, CHLORIDE ION, Dihydrofolate Reductase, ... | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-10-22 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Propargyl-Linked Antifolates are Dual Inhibitors of Candida albicans and Candida glabrata.
J.Med.Chem., 57, 2014
|
|
4HOF
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-[3-(2-methoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine (UCP111H) | | Descriptor: | 5-[3-(2-methoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-10-22 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Propargyl-Linked Antifolates are Dual Inhibitors of Candida albicans and Candida glabrata.
J.Med.Chem., 57, 2014
|
|
4HOE
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine (UCP111E) | | Descriptor: | 5-[3-(2,5-dimethoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-10-22 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Propargyl-Linked Antifolates are Dual Inhibitors of Candida albicans and Candida glabrata.
J.Med.Chem., 57, 2014
|
|
4BEM
 
 | | Crystal structure of the F-type ATP synthase c-ring from Acetobacterium woodii. | | Descriptor: | ACETATE ION, F1FO ATPASE C1 SUBUNIT, F1FO ATPASE C2 SUBUNIT, ... | | Authors: | Matthies, D, Meier, T, Yildiz, O. | | Deposit date: | 2013-03-11 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Resolution Structure and Mechanism of an F/V-Hybrid Rotor Ring in a Na+-Coupled ATP Synthase
Nat.Commun., 5, 2014
|
|
4IF2
 
 | |
6H86
 
 | |
4AGW
 
 | | Discovery of a small molecule type II inhibitor of wild-type and gatekeeper mutants of BCR-ABL, PDGFRalpha, Kit, and Src kinases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-{2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromet hyl)phenyl}benzamide, GLYCEROL, ... | | Authors: | Weisberg, E, Choi, H.G, Seeliger, M, Gray, N, Griffin, J.D. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Small-Molecule Type II Inhibitor of Wild-Type and Gatekeeper Mutants of Bcr-Abl, Pdgfralpha, Kit, and Src Kinases: Novel Type II Inhibitor of Gatekeeper Mutants.
Blood, 115, 2010
|
|
3PDV
 
 | |
2XKY
 
 | | Single particle analysis of Kir2.1NC_4 in negative stain | | Descriptor: | INWARD RECTIFIER POTASSIUM CHANNEL 2 | | Authors: | Fomina, S, Howard, T.D, Sleator, O.K, Golovanova, M, O'Ryan, L, Leyland, M.L, Grossmann, J.G, Collins, R.F, Prince, S.M. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17.200001 Å), SOLUTION SCATTERING | | Cite: | Self-Directed Assembly and Clustering of the Cytoplasmic Domains of Inwardly Rectifying Kir2.1 Potassium Channels on Association with Psd-95
Biochim.Biophys.Acta, 1808, 2011
|
|
4PZA
 
 | | The complex structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c with inorganic phosphate | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase, PHOSPHATE ION | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-03-29 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
4PZ9
 
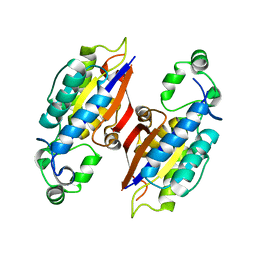 | | The native structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-03-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
4QIH
 
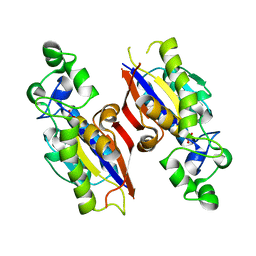 | | The structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c complexes with VO3 | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase, VANADATE ION | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
4RVS
 
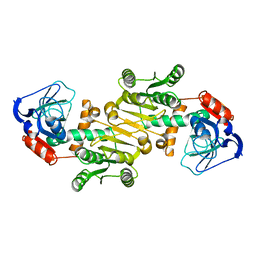 | | The native structure of mycobacterial quinone oxidoreductase Rv154c. | | Descriptor: | Probable quinone reductase Qor (NADPH:quinone reductase) (Zeta-crystallin homolog protein) | | Authors: | Zhou, W.H, Zheng, Q.Q, Song, Y.L, Zhang, W, Shaw, N, Rao, Z. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8464 Å) | | Cite: | Structural views of quinone oxidoreductase from Mycobacterium tuberculosis reveal large conformational changes induced by the co-factor.
Febs J., 282, 2015
|
|
4RVU
 
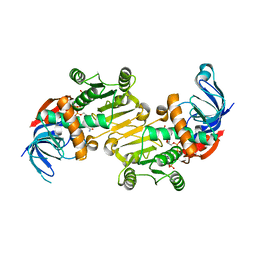 | | The native structure of mycobacterial Rv1454c complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable quinone reductase Qor (NADPH:quinone reductase) (Zeta-crystallin homolog protein) | | Authors: | Zhou, W.H, Zheng, Q.Q, Song, Y.L, Zhang, W, Shaw, N, Rao, Z. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7988 Å) | | Cite: | Structural views of quinone oxidoreductase from Mycobacterium tuberculosis reveal large conformational changes induced by the co-factor.
Febs J., 282, 2015
|
|
1GIS
 
 | | A TRICHOSANTHIN(TCS) MUTANT(E85Q) COMPLEX STRUCTURE WITH 2'-DEOXY-ADENOSIN-5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Guo, Q, Liu, Y, Dong, Y, Rao, Z. | | Deposit date: | 2001-03-15 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate binding and catalysis in trichosanthin occur in different sites as revealed by the complex structures of several E85 mutants.
Protein Eng., 16, 2003
|
|
2G2X
 
 | | X-Ray Crystal Structure Protein Q88CH6 from Pseudomonas putida. Northeast Structural Genomics Consortium Target PpR72. | | Descriptor: | SULFATE ION, hypothetical protein PP5205 | | Authors: | Kuzin, A.P, Chen, Y, Abashidze, M, Acton, T, Conover, K, Janjua, H, Ma, L.-C, Ho, C.K, Cunningham, K, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-02-16 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
