7RDH
 
 | |
4FIH
 
 | |
4FIJ
 
 | | Catalytic domain of human PAK4 | | Descriptor: | Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FIE
 
 | | Full-length human PAK4 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FIG
 
 | | Catalytic domain of human PAK4 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FIF
 
 | | Catalytic domain of human PAK4 with RPKPLVDP peptide | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GZM
 
 | | Crystal structure of RAC1 F28L mutant | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-09-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RAC1P29S is a spontaneously activating cancer-associated GTPase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GZL
 
 | | Crystal structure of RAC1 Q61L mutant | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-09-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RAC1P29S is a spontaneously activating cancer-associated GTPase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1X11
 
 | | X11 PTB DOMAIN | | Descriptor: | 13-MER PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
7N1J
 
 | |
7N1K
 
 | |
7N3T
 
 | | TrkA ECD complex with designed miniprotein ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Cao, L, Garcia, K.C. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
1QG1
 
 | |
3SBE
 
 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
1GCQ
 
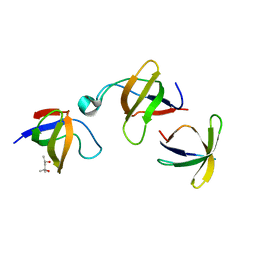 | | CRYSTAL STRUCTURE OF VAV AND GRB2 SH3 DOMAINS | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
3SBD
 
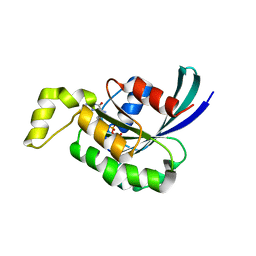 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
3TH5
 
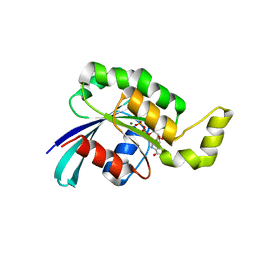 | | Crystal structure of wild-type RAC1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
1GCP
 
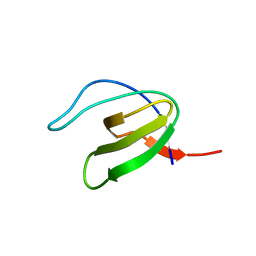 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1PBW
 
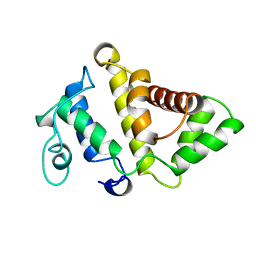 | | STRUCTURE OF BCR-HOMOLOGY (BH) DOMAIN | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Musacchio, A, Cantley, L.C, Harrison, S.C. | | Deposit date: | 1996-10-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the breakpoint cluster region-homology domain from phosphoinositide 3-kinase p85 alpha subunit.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1GFD
 
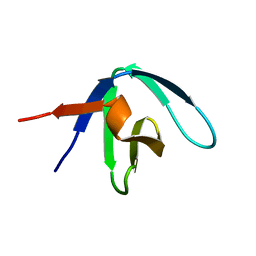 | |
1QQL
 
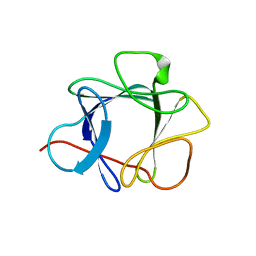 | | THE CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 7/1 CHIMERA | | Descriptor: | Fibroblast growth factor 7, Fibroblast growth factor 1 chimera | | Authors: | Ye, S, Luo, Y, Pelletier, H, McKeehan, W.L. | | Deposit date: | 1999-06-07 | | Release date: | 2000-01-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs.
Biochemistry, 40, 2001
|
|
1QQK
 
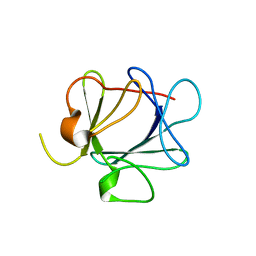 | | THE CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 7 (KERATINOCYTE GROWTH FACTOR) | | Descriptor: | FIBROBLAST GROWTH FACTOR 7 | | Authors: | Ye, S, Luo, Y, Pelletier, H, McKeehan, W.L. | | Deposit date: | 1999-06-07 | | Release date: | 2000-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs.
Biochemistry, 40, 2001
|
|
1GFC
 
 | |
1H5P
 
 | | Solution structure of the human Sp100b SAND domain by heteronuclear NMR. | | Descriptor: | NUCLEAR AUTOANTIGEN SP100-B | | Authors: | Bottomley, M.J, Liu, Z, Collard, M.W, Huggenvik, J.I, Gibson, T.J, Sattler, M. | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation.
Nat. Struct. Biol., 8, 2001
|
|
