6K4J
 
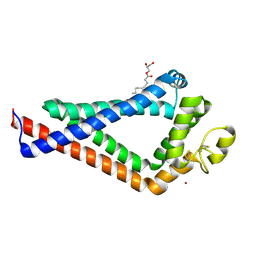 | | Crystal Structure of the the CD9 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CD9 antigen, NICKEL (II) ION, ... | | Authors: | Umeda, R, Nishizawa, T, Sato, K, Nureki, O. | | Deposit date: | 2019-05-24 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural insights into tetraspanin CD9 function.
Nat Commun, 11, 2020
|
|
8WCI
 
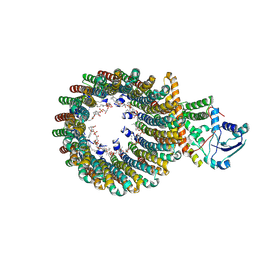 | | Cryo-EM structure of the inhibitor-bound Vo complex from Enterococcus hirae | | Descriptor: | CARDIOLIPIN, N,N-dimethyl-4-(5-methyl-1H-benzimidazol-2-yl)aniline, SODIUM ION, ... | | Authors: | Suzuki, K, Mikuriya, S, Adachi, N, Kawasaki, M, Senda, T, Moriya, T, Murata, T. | | Deposit date: | 2023-09-12 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Novel Inhibitor of Na+-Transporting V-ATPase Suppresses VRE Colonization in Mice and Reveals the High-Resolution Structure of the Na+ Transport Pathway
To Be Published
|
|
2FZA
 
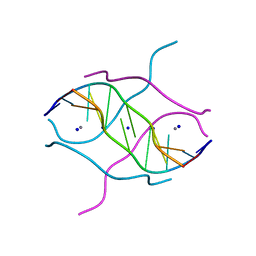 | | Crystal structure of d(GCGGGAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*GP*GP*AP*GP*C)-3', CALCIUM ION, SODIUM ION | | Authors: | Kondo, J, Ciengshin, T, Juan, E.C.M, Mitomi, K, Takenaka, A. | | Deposit date: | 2006-02-09 | | Release date: | 2007-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of d(gcGXGAgc) with X=G: a mutation at X is possible to occur in a base-intercalated duplex for multiplex formation
Nucleosides Nucleotides Nucleic Acids, 25, 2006
|
|
7KVZ
 
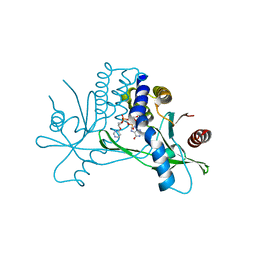 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-2 | | Descriptor: | (2R,5R,7R,8R,10S,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2,10,16-trihydroxy-14-[(pyrimidin-4-yl)oxy]decahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R.J. | | Deposit date: | 2020-11-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KW1
 
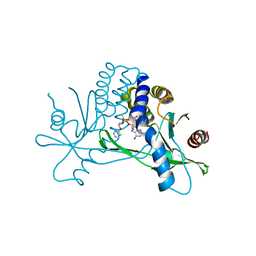 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-3 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)oxy]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVX
 
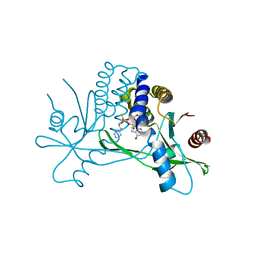 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN 1 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)amino]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
4YB9
 
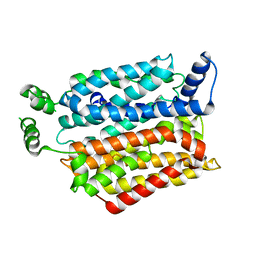 | | Crystal structure of the Bovine Fructose transporter GLUT5 in an open inward-facing conformation | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5 | | Authors: | Verdon, G, Kang, H.J, Iwata, S, Drew, D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5.
Nature, 526, 2015
|
|
4YBQ
 
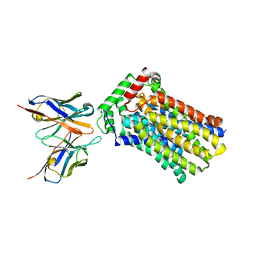 | | Rat GLUT5 with Fv in the outward-open form | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5, antibody Fv fragment heavy chain, ... | | Authors: | Nomura, N, Shimamura, T, Iwata, S. | | Deposit date: | 2015-02-19 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5
Nature, 526, 2015
|
|
5GTF
 
 | | Crystal structure of onion lachrymatory factor synthase (LFS) containing glycerol | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lachrymatory-factor synthase, ... | | Authors: | Takabe, J, Arakawa, T, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2016-08-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase
Acs Catalysis, 10, 2020
|
|
5GTE
 
 | | Crystal structure of onion lachrymatory factor synthase (LFS), solute-free form | | Descriptor: | Lachrymatory-factor synthase, SULFATE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Takabe, J, Arakawa, T, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2016-08-20 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase
Acs Catalysis, 10, 2020
|
|
5B4O
 
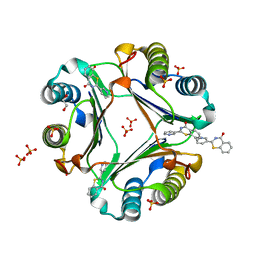 | | Crystal structure of Macrophage Migration Inhibitory Factor in complex with BTZO-14 | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-1,3-benzothiazin-4-one, Macrophage migration inhibitory factor, ... | | Authors: | Oki, H, Igaki, S, Moriya, Y, Hayano, Y, Habuka, N. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression
Chem. Biol., 17, 2010
|
|
8OOI
 
 | | Full composite cryo-EM map of p97/VCP in ADP.Pi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cheng, T.C, Sakata, E, Schuetz, A.K. | | Deposit date: | 2023-04-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Characterizing ATP processing by the AAA+ protein p97 at the atomic level.
Nat.Chem., 16, 2024
|
|
7VAF
 
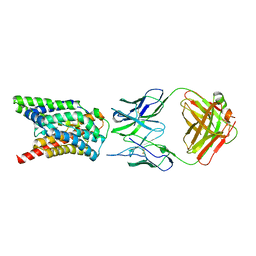 | | Cryo-EM structure of Rat NTCP complexed with YN69202Fab | | Descriptor: | Fab heavy chain from antibody IgG clone number YN69202, Fab light chain from antibody IgG clone number YN69202, Sodium/bile acid cotransporter | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2021-08-29 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of the bile acid transporter and HBV receptor NTCP.
Nature, 606, 2022
|
|
7VAG
 
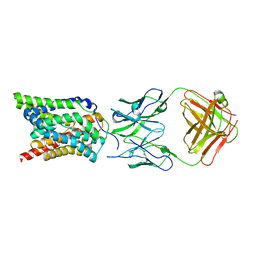 | |
7VAD
 
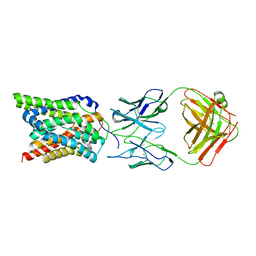 | | Cryo-EM structure of human NTCP complexed with YN69202Fab | | Descriptor: | Fab heavy chain from antibody IgG clone number YN69202, Fab light chain from antibody IgG clone number YN69202, Sodium/bile acid cotransporter | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2021-08-29 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the bile acid transporter and HBV receptor NTCP.
Nature, 606, 2022
|
|
7VAE
 
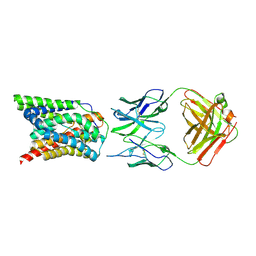 | | Cryo-EM structure of bovine NTCP complexed with YN69202Fab | | Descriptor: | Fab heavy chain from antibody IgG clone number YN69202, Fab light chain from antibody IgG clone number YN69202, Solute carrier family 10 (Sodium/bile acid cotransporter family), ... | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2021-08-29 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the bile acid transporter and HBV receptor NTCP.
Nature, 606, 2022
|
|
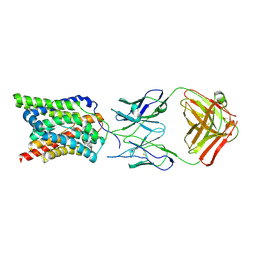
7WSI
 
 | |
5B8C
 
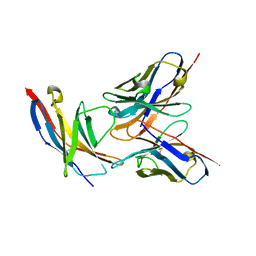 | | High resolution structure of the human PD-1 in complex with pembrolizumab Fv | | Descriptor: | Pembrolizumab heavy chain variable region (PemVH), Pembrolizumab light chain variable region (PemVL), Programmed cell death protein 1 | | Authors: | Horita, S, Shimamura, T, Iwata, S, Nomura, N. | | Deposit date: | 2016-06-14 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1
Sci Rep, 6, 2016
|
|
8HRX
 
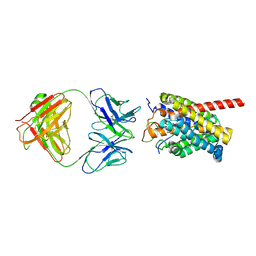 | | Cryo-EM structure of human NTCP-myr-preS1-YN9048Fab complex | | Descriptor: | Fab heavy chain from antibody IgG clone number YN9048, Fab light chain from antibody IgG clone number YN9048, PreS1 protein (Fragment), ... | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis of hepatitis B virus receptor binding.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HRY
 
 | | Cryo-EM structure of human NTCP-myr-preS1-YN9016Fab complex | | Descriptor: | Fab heavy chain from antibody IgG clone number YN9016, Fab light chain from antibody IgG clone number YN9016, Large S protein (Fragment), ... | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hepatitis B virus receptor binding.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5H11
 
 | |
7DC0
 
 | |
7DC4
 
 | | Crystal structure of glycan-bound Pseudomonas taiwanensis lectin | | Descriptor: | Lectin, SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lectins engineered to favor a glycan-binding conformation have enhanced antiviral activity.
J.Biol.Chem., 296, 2021
|
|
4YH7
 
 | | Crystal structure of PTPdelta ectodomain in complex with IL1RAPL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
4YFE
 
 | | Crystal structure of PTP delta Fn1-Fn2 | | Descriptor: | Receptor-type tyrosine-protein phosphatase delta | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
