6QXX
 
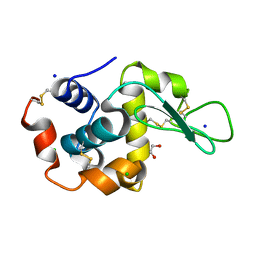 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 14793 patterns merged | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
5J7A
 
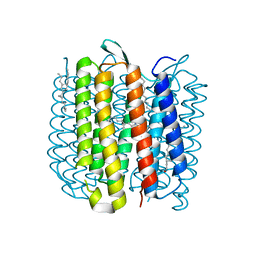 | | Bacteriorhodopsin ground state structure obtained with Serial Femtosecond Crystallography | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Panneels, V, Nelson, G, Gati, C, Kimura, T, Milne, C, Milathianaki, D, Kubo, M, Wu, W, Conrad, C, Coe, J, Bean, R, Zhao, Y, Bath, P, Dods, R, Harimoorthy, R, Beyerlein, K.R, Rheinberger, J, James, D, DePonte, D, Li, C, Sala, L, Williams, G, Hunter, M, Koglin, J.E, Berntsen, P, Nango, E, Iwata, S, Chapman, H.N, Fromme, P, Frank, M, Abela, R, Boutet, S, Barty, A, White, T.A, Weierstall, U, Spence, J, Neutze, R, Schertler, G, Standfuss, J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography.
Nat Commun, 7, 2016
|
|
5HD3
 
 | |
6U57
 
 | |
5HDC
 
 | |
5HDD
 
 | |
5HDS
 
 | |
5HD5
 
 | |
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4LQY
 
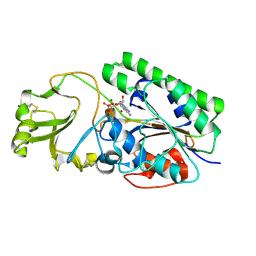 | | Crystal Structure of Human ENPP4 with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Albright, R.A, Braddock, D.T. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular basis of purinergic signal metabolism by ectonucleotide pyrophosphatase/phosphodiesterases 4 and 1 and implications in stroke.
J.Biol.Chem., 289, 2014
|
|
4LR2
 
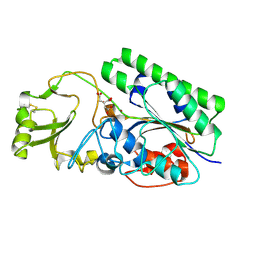 | | Crystal Structure of Human ENPP4 (apo) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bis(5'-adenosyl)-triphosphatase ENPP4, ... | | Authors: | Albright, R.A, Braddock, D.T. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of purinergic signal metabolism by ectonucleotide pyrophosphatase/phosphodiesterases 4 and 1 and implications in stroke.
J.Biol.Chem., 289, 2014
|
|
5G3X
 
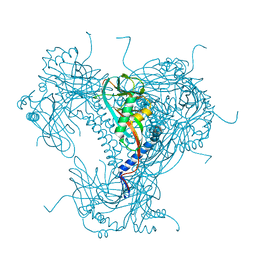 | | Structure of recombinant granulovirus polyhedrin | | Descriptor: | GRANULOVIRUS POLYHEDRIN | | Authors: | Bunker, R.D, Chiu, E, Metcalf, P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6B69
 
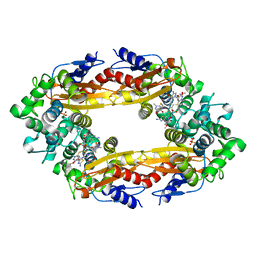 | | Beta-Lactamase, 500ms timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, (2R)-2-[(S)-{[(2E)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-(hydroxymethyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6D
 
 | | Beta-Lactamase, mixed with Ceftriaxone, needles crystal form, 100ms | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B5Y
 
 | | Beta-lactamase, mixed with Ceftriaxone, 30ms time point, Shards crystal form | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6B
 
 | |
6B6A
 
 | | Beta-Lactamase, 2secs timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6C
 
 | |
6B6E
 
 | | Beta-Lactamase, mixed with Ceftriaxone, needles crystal form, 500ms | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B68
 
 | | Beta-Lactamase, 100ms timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B5X
 
 | | Beta-Lactamase, unmixed shards crystal form | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Pandey, S. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6F
 
 | |
7Z2K
 
 | | Crystal structure of SARS-CoV-2 Main Protease in orthorhombic space group p212121 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-02-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
6VWV
 
 | |
6VWT
 
 | |
