3DJU
 
 | | Crystal structure of human BTG2 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
3OFU
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
3OFT
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (2R,5R)-hexane-2,5-diol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
3NGO
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with poly(A) DNA | | Descriptor: | 5'-D(*AP*AP*AP*A)-3', CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
2X81
 
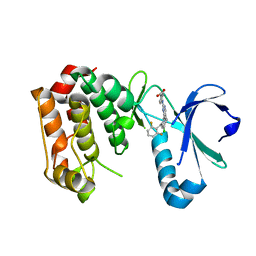 | | STRUCTURE OF AURORA A IN COMPLEX WITH MLN8054 | | Descriptor: | 4-{[9-CHLORO-7-(2,6-DIFLUOROPHENYL)-5H-PYRIMIDO[5,4-D][2]BENZAZEPIN-2-YL]AMINO}BENZOIC ACID, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Savory, W, Mueller, I, Mason, C.S, Lamers, M, Williams, D.H, Eyers, P.A. | | Deposit date: | 2010-03-05 | | Release date: | 2010-05-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Drug-Resistant Aurora a Mutants for Cellular Target Validation of the Small Molecule Kinase Inhibitors Mln8054 and Mln8237.
Acs Chem.Biol., 5, 2010
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6D
 
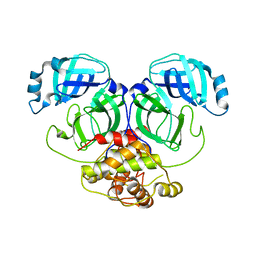 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | Descriptor: | Infectious bronchitis virus (IBV) main protease | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
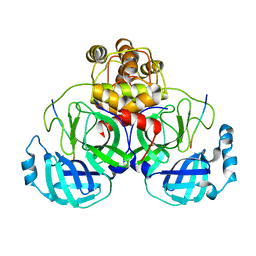 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | Descriptor: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2QC3
 
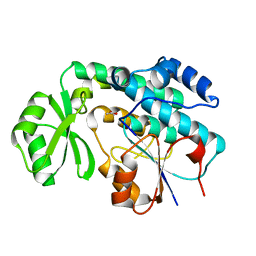 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
6IN8
 
 | | Crystal structure of MucB | | Descriptor: | Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6IKJ
 
 | | Crystal structure of YfiB(F48S) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IN9
 
 | | Crystal structure of MucB in complex with MucA(peri) | | Descriptor: | Sigma factor AlgU negative regulatory protein, Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6INB
 
 | |
6INC
 
 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6IN7
 
 | | Crystal structure of AlgU in complex with MucA(cyto) | | Descriptor: | NICOTINAMIDE, RNA polymerase sigma-H factor, Sigma factor AlgU negative regulatory protein | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
7PKR
 
 | | Vault structure in primmed conformation | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7PKZ
 
 | | Vault structure in committed conformation | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7PKY
 
 | | Half-vault structure | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
6JC4
 
 | |
8BYA
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
6JYV
 
 | | Structure of an isopenicillin N synthase from Pseudomonas aeruginosa PAO1 | | Descriptor: | Probable iron/ascorbate oxidoreductase, SODIUM ION | | Authors: | Hao, Z, Che, S, Wang, R, Liu, R, Zhang, Q, Bartlam, M. | | Deposit date: | 2019-04-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural characterization of an isopenicillin N synthase family oxygenase from Pseudomonas aeruginosa PAO1.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
6IKI
 
 | | Crystal structure of YfiB(W55L) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IKK
 
 | |
7QE8
 
 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 (SPINK1) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
7QE9
 
 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 N34S (SPINK1 N34S) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
