4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1UGY
 
 | | Crystal structure of jacalin- mellibiose (Gal-alpha(1-6)-Glc) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1UGX
 
 | | Crystal structure of jacalin- Me-alpha-T-antigen (Gal-beta(1-3)-GalNAc-alpha-o-Me) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1VBP
 
 | | Crystal structure of artocarpin-mannopentose complex | | Descriptor: | SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Jeyaprakash, A.A, Srivastav, A, Surolia, A, Vijayan, M. | | Deposit date: | 2004-02-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity
J.Mol.Biol., 338, 2004
|
|
1VBO
 
 | | Crystal structure of artocarpin-mannotriose complex | | Descriptor: | alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, artocarpin | | Authors: | Jeyaprakash, A.A, Srivastav, A, Surolia, A, Vijayan, M. | | Deposit date: | 2004-02-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity
J.Mol.Biol., 338, 2004
|
|
4C11
 
 | |
5NYA
 
 | | Carbonic Anhydrase II Inhibitor RA13 | | Descriptor: | Carbonic anhydrase 2, GLUTARIC ACID, ZINC ION, ... | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NY1
 
 | | Carbonic Anhydrase II Inhibitor RA10 | | Descriptor: | 3,6,7-trimethyl-~{N}-[(4-sulfamoylphenyl)methyl]-1-benzofuran-2-carboxamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NY6
 
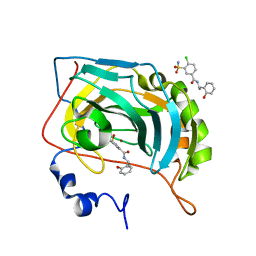 | | Carbonic Anhydrase II Inhibitor RA12 | | Descriptor: | 4-chloranyl-~{N}-[(1~{R})-1-(2-hydroxyphenyl)ethyl]-3-sulfamoyl-benzamide, 4-chloranyl-~{N}-[(1~{S})-1-(2-hydroxyphenyl)ethyl]-3-sulfamoyl-benzamide, Carbonic anhydrase 2, ... | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXG
 
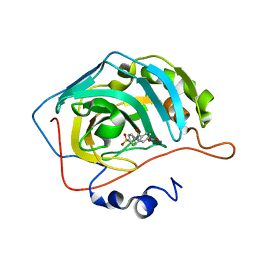 | | Carbonic Anhydrase II Inhibitor RA1 | | Descriptor: | 2-chloranyl-4-nitro-~{N}-(4-sulfamoylphenyl)benzamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXP
 
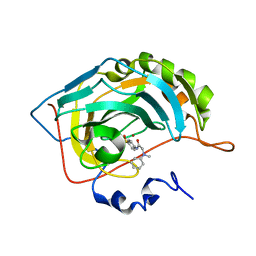 | | Carbonic Anhydrase II Inhibitor RA7 | | Descriptor: | 2-[methyl(phenyl)amino]-~{N}-(4-sulfamoylphenyl)ethanamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXW
 
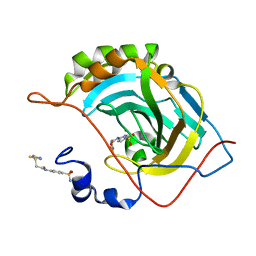 | | Carbonic Anhydrase II Inhibitor RA9 | | Descriptor: | 2-[(5-azanyl-1,3,4-thiadiazol-2-yl)sulfanyl]-~{N}-(4-sulfamoylphenyl)ethanamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXM
 
 | | Carbonic Anhydrase II Inhibitor RA1 | | Descriptor: | 8-methoxy-2-oxidanylidene-~{N}-(4-sulfamoylphenyl)chromene-3-carboxamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXI
 
 | | Carbonic Anhydrase II Inhibitor RA2 | | Descriptor: | 4-[(4-bromophenyl)sulfonylamino]benzenesulfonamide, CITRIC ACID, Carbonic anhydrase 2, ... | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXO
 
 | | Carbonic Anhydrase II Inhibitor RA6 | | Descriptor: | 4-[(4-nitrophenyl)methyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXV
 
 | | Carbonic Anhydrase II Inhibitor RA8 | | Descriptor: | 2-(4-chloranyl-5-methyl-3-nitro-pyrazol-1-yl)-~{N}-(4-sulfamoylphenyl)ethanamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NY3
 
 | | Carbonic Anhydrase II Inhibitor RA11 | | Descriptor: | 1-(4-chlorophenyl)-3-[2-(4-sulfamoylphenyl)ethyl]urea, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
2D3S
 
 | | Crystal Structure of basic winged bean lectin with Tn-antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, ... | | Authors: | Kulkarni, K.A, Sinha, S, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2005-10-01 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the specificity of basic winged bean lectin for the Tn-antigen: a crystallographic, thermodynamic and modelling study
Febs Lett., 579, 2005
|
|
2DU1
 
 | | Crystal Structure of basic winged bean lectin in complex with Methyl-alpha-N-acetyl-D galactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2006-07-19 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the carbohydrate-specificity of basic winged-bean lectin and its differential affinity for Gal and GalNAc
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DTY
 
 | | Crystal structure of basic winged bean lectin complexed with N-acetyl-D-galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2006-07-19 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the carbohydrate-specificity of basic winged-bean lectin and its differential affinity for Gal and GalNAc
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
4KBA
 
 | | CK1d in complex with 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine inhibitor | | Descriptor: | 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine, Casein kinase I isoform delta, SULFATE ION | | Authors: | Liu, S. | | Deposit date: | 2013-04-23 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Ligand-protein interactions of selective casein kinase 1 delta inhibitors.
J.Med.Chem., 56, 2013
|
|
4KBK
 
 | |
2E7T
 
 | | Crystal structure of basic winged bean lectin in complex with a blood group trisaccharide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2007-01-13 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Generation of blood group specificity: New insights from structural studies on the complexes of A- and B-reactive saccharides with basic winged bean agglutinin
Proteins, 68, 2007
|
|
2E53
 
 | | Crystal structure of basic winged bean lectin in complex with B blood group disaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2006-12-18 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Generation of blood group specificity: new insights from structural studies on the complexes of A- and B-reactive saccharides with basic winged bean agglutinin.
Proteins, 68, 2007
|
|
