6KQT
 
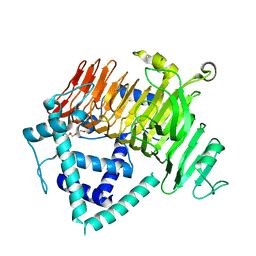 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein | | Descriptor: | SODIUM ION, TRIETHYLENE GLYCOL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
6KQS
 
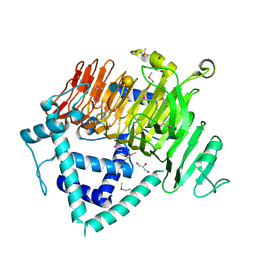 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lacto-N-biosidase | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
1Z6H
 
 | | Solution Structure of Bacillus subtilis BLAP biotinylated-form | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Biotin/Lipoyl Attachment Protein | | Authors: | Cui, G, Xia, B. | | Deposit date: | 2005-03-22 | | Release date: | 2006-03-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Identification and solution structures of a single domain biotin/lipoyl attachment protein from Bacillus subtilis
J.Biol.Chem., 281, 2006
|
|
2LIZ
 
 | |
2LKT
 
 | |
2MY9
 
 | |
2KYT
 
 | | Solution structure of the H-REV107 N-terminal domain | | Descriptor: | Group XVI phospholipase A2 | | Authors: | Ren, X, Xia, B. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal catalytic domain of human H-REV107--a novel circular permutated NlpC/P60 domain
Febs Lett., 584, 2010
|
|
5YRZ
 
 | | Toxin-Antitoxin complex from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, HicA, HicB, ... | | Authors: | Kang, S.M, Kim, D.H. | | Deposit date: | 2017-11-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Functional insights into the Streptococcus pneumoniae HicBA toxin-antitoxin system based on a structural study.
Nucleic Acids Res., 46, 2018
|
|
8YZG
 
 | | Antitoxin protein from S. pneumoniae | | Descriptor: | Antitoxin | | Authors: | Kim, D.H, Kang, S.M, Lee, B.J. | | Deposit date: | 2024-04-06 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Insight Into YefM-YoeB Complex of Toxin-Antitoxin System From Streptococcus pneumoniae.
J.Cell.Biochem., 126, 2025
|
|
6A4W
 
 | | AcrR from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Kang, S.M, Kim, D.H. | | Deposit date: | 2018-06-21 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | The crystal structure of AcrR from Mycobacterium tuberculosis reveals a one-component transcriptional regulation mechanism.
Febs Open Bio, 9, 2019
|
|
6A4L
 
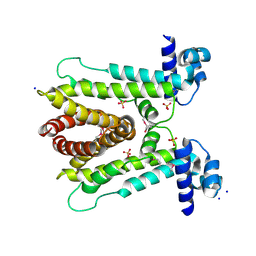 | | AcrR from Mycobacterium tuberculosis | | Descriptor: | SODIUM ION, SULFATE ION, TetR family transcriptional regulator | | Authors: | Kang, S.M. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of AcrR from Mycobacterium tuberculosis reveals a one-component transcriptional regulation mechanism.
Febs Open Bio, 9, 2019
|
|
8XLK
 
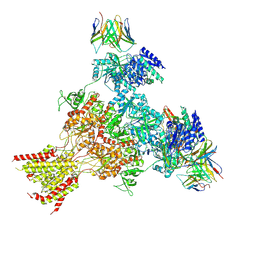 | | Structure of native tri-heteromeric GluN1-GluN2A-GluN2B NMDA receptor in rat cortex and hippocampus | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, M, Feng, J, Li, Y, Zhu, S. | | Deposit date: | 2023-12-26 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Assembly and architecture of endogenous NMDA receptors in adult cerebral cortex and hippocampus.
Cell, 188, 2025
|
|
8XLL
 
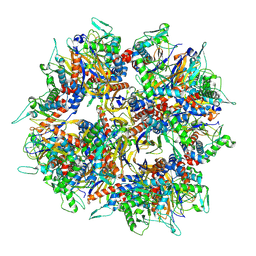 | | Structure of the native 2-oxoglutarate dehydrogenase complex (OGDHC) in the adult cortex and hippocampus | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Zhang, M, Feng, J, Li, Y, Zhu, S. | | Deposit date: | 2023-12-26 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Assembly and architecture of endogenous NMDA receptors in adult cerebral cortex and hippocampus.
Cell, 188, 2025
|
|
8XLJ
 
 | |
7U34
 
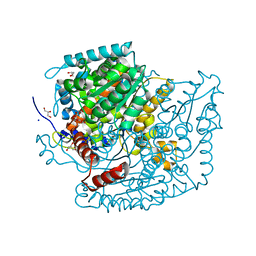 | | The structure of phosphoglucose isomerase from Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Yan, K, Kowalski, B, Fang, W, van Aalten, D. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
7DFB
 
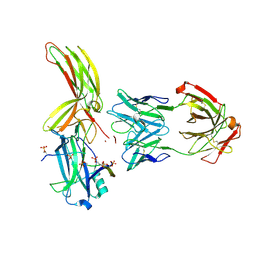 | | Crystal of Arrestin2-V2Rpp-6-7-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DF9
 
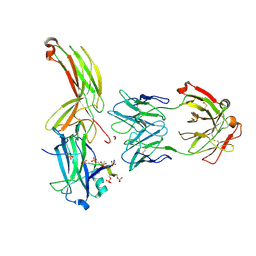 | | Crystal of Arrestin2-V2Rpp-1-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFA
 
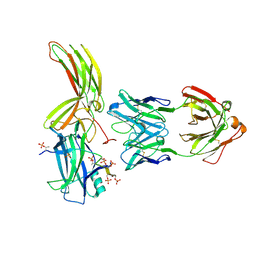 | | Crystal of Arrestin2-V2Rpp-4-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFC
 
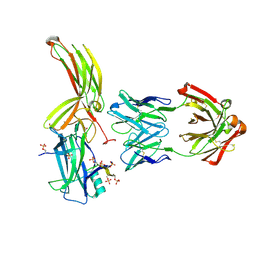 | | Crystal of Arrestin2-V2Rpp-3-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
6JMQ
 
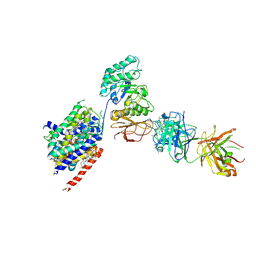 | | LAT1-CD98hc complex bound to MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Nakane, T, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8K31
 
 | | The complex of WRKY33 C terminal DBD and SIB1 | | Descriptor: | Probable WRKY transcription factor 33, Sigma factor binding protein 1, chloroplastic, ... | | Authors: | Dong, X, Gong, Z, Hu, Y.F. | | Deposit date: | 2023-07-14 | | Release date: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1.
Commun Biol, 7, 2024
|
|
6JMR
 
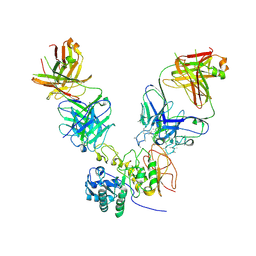 | | CD98hc extracellular domain bound to HBJ127 Fab and MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Yokoyama, T, Nakane, T, Shirouzu, M, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7E6H
 
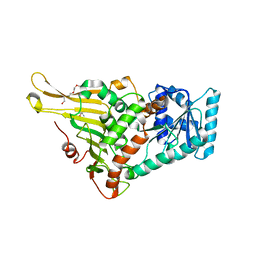 | | glucose-6-phosphate dehydrogenase from Kluyveromyces lactis | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Ha, V.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
7E6I
 
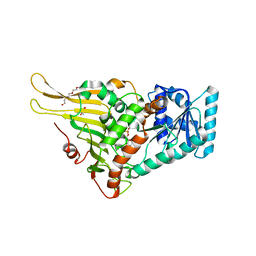 | | Glucose-6-phosphate dehydrogenase in complex with its substrate glucose-6-phosphate | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Vu, H.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
8KDN
 
 | | Structure of LAT1-CD98hc in complex with L-Phe, focused on TMD | | Descriptor: | 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1, PHENYLALANINE | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
