1LO8
 
 | | X-ray crystal structure of 4-hydroxybenzoyl CoA thioesterase complexed with 4-hydroxybenzyl CoA | | Descriptor: | 4-HYDROXYBENZYL COENZYME A, 4-hydroxybenzoyl-CoA Thioesterase | | Authors: | Thoden, J.B, Holden, H.M, Zhuang, Z, Dunaway-Mariano, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase.
J.Biol.Chem., 277, 2002
|
|
3M9V
 
 | |
3MQH
 
 | | crystal structure of the 3-N-acetyl transferase WlbB from Bordetella petrii in complex with CoA and UDP-3-amino-2-acetamido-2,3-dideoxy glucuronic acid | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | thoden, J.B, holden, H.M. | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular structure of WlbB, a bacterial N-acetyltransferase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid .
Biochemistry, 49, 2010
|
|
3MQG
 
 | | crystal structure of the 3-N-acetyl transferase WlbB from Bordetella petrii in complex with acetyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETYL COENZYME *A, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular structure of WlbB, a bacterial N-acetyltransferase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid .
Biochemistry, 49, 2010
|
|
3NDJ
 
 | | X-ray Structure of a C-3'-Methyltransferase in Complex with S-Adenosyl-L-Homocysteine and Sugar Product | | Descriptor: | (2R,4S,6R)-4-amino-4,6-dimethyl-5-oxotetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), Methyltransferase, PHOSPHATE ION, ... | | Authors: | Bruender, N.A, Thoden, J.B, Kaur, M, Avey, M.K, Holden, H.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Architecture of a C-3'-Methyltransferase Involved in the Biosynthesis of d-Tetronitrose.
Biochemistry, 49, 2010
|
|
3NDI
 
 | | X-ray Structure of a C-3'-Methyltransferase in Complex with S-adenosylmethionine and dTMP | | Descriptor: | Methyltransferase, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Bruender, N.A, Thoden, J.B, Kaur, M, Avey, M.K, Holden, H.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Architecture of a C-3'-Methyltransferase Involved in the Biosynthesis of d-Tetronitrose.
Biochemistry, 49, 2010
|
|
4I3G
 
 | | Crystal Structure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Beta-glucosidase, ... | | Authors: | Zmudka, M.W, Holden, H.M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of DesR from Streptomyces venezuelae, a beta-glucosidase involved in macrolide activation.
Protein Sci., 22, 2013
|
|
3B8X
 
 | | Crystal structure of GDP-4-keto-6-deoxymannose-3-dehydratase (ColD) H188N mutant with bound GDP-perosamine | | Descriptor: | 1,2-ETHANEDIOL, Pyridoxamine 5-phosphate-dependent dehydrase, SODIUM ION, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GDP-4-Keto-6-deoxy-D-mannose 3-Dehydratase, Accommodating a Sugar Substrate in the Active Site.
J.Biol.Chem., 283, 2008
|
|
3BN1
 
 | | Crystal structure of GDP-perosamine synthase | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, Perosamine synthetase, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-12-13 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GDP-Perosamine Synthase: Structural Analysis and Production of a Novel Trideoxysugar
Biochemistry, 47, 2008
|
|
6Q03
 
 | | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine) | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine)
To Be Published
|
|
6Q0A
 
 | | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid
To Be Published
|
|
6Q11
 
 | | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID
To Be Published
|
|
6PZ2
 
 | |
6Q0Y
 
 | | Crystal structure of MurA from Clostridium difficile, mutant C116S, in the presence of Uridine-Diphosphate-N-Acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutant C116S, in the presence of Uridine-Diphosphate-N-Acetylglucosamine
To Be Published
|
|
6OFU
 
 | | X-ray crystal structure of the YdjI aldolase from Escherichia coli K12 | | Descriptor: | CHLORIDE ION, YdjI aldolase, ZINC ION | | Authors: | Dopkins, B.J, Thoden, J.B, Huddleston, J.P, Narindoshvili, T, Fose, B, Rachel, F.M, Holden, H.M. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Characterization of YdjI, an Aldolase of Unknown Specificity inEscherichia coliK12.
Biochemistry, 58, 2019
|
|
2GMU
 
 | | Crystal structure of E coli GDP-4-keto-6-deoxy-D-mannose-3-dehydratase complexed with PLP-glutamate ketimine intermediate | | Descriptor: | MAGNESIUM ION, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID, Putative pyridoxamine 5-phosphate-dependent dehydrase, ... | | Authors: | Cook, P.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme.
Protein Sci., 15, 2006
|
|
2GMS
 
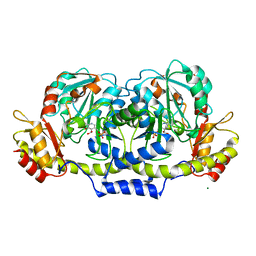 | | E coli GDP-4-keto-6-deoxy-D-mannose-3-dehydratase with bound hydrated PLP | | Descriptor: | MAGNESIUM ION, Putative pyridoxamine 5-phosphate-dependent dehydrase, Wbdk, ... | | Authors: | Cook, P.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme.
Protein Sci., 15, 2006
|
|
1BXK
 
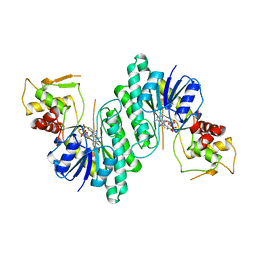 | | DTDP-GLUCOSE 4,6-DEHYDRATASE FROM E. COLI | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (DTDP-GLUCOSE 4,6-DEHYDRATASE) | | Authors: | Thoden, J.B, Hegeman, A.D, Frey, P.A, Holden, H.M. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Structure of Dtdp-Glucose 4,6-Dehydratase from E. Coli
Protein Sci.
|
|
2E3D
 
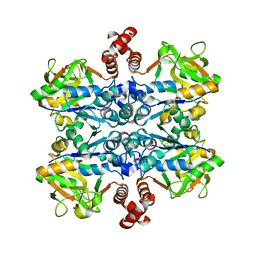 | |
3E1K
 
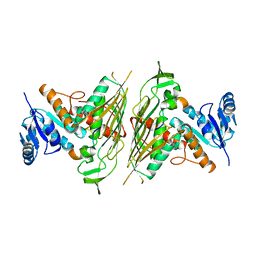 | |
3FRK
 
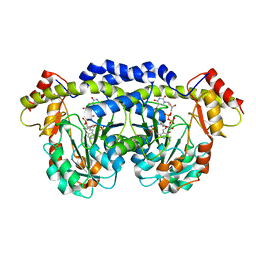 | | X-ray structure of QdtB from T. thermosaccharolyticum in complex with a PLP:TDP-3-aminoquinovose aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, QdtB | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of QdtB, an aminotransferase required for the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose.
Biochemistry, 48, 2009
|
|
2UDP
 
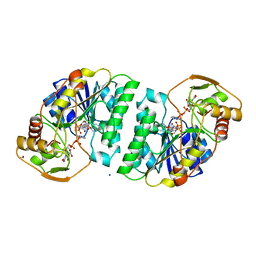 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHENYL-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol.
Protein Sci., 5, 1996
|
|
6V2T
 
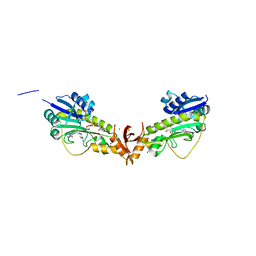 | | X-ray structure of a sugar N-formyltransferase from Shewanella sp FDAARGOS_354 | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, PHOSPHATE ION, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
6VO6
 
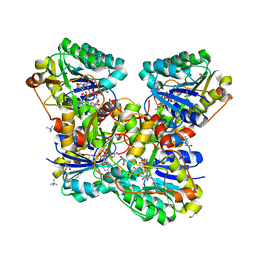 | | Crystal Structure of Cj1427, an Essential NAD-dependent Dehydrogenase from Campylobacter jejuni, in the Presence of NADH and GDP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Anderson, T.K, Spencer, K.D, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6VO8
 
 | | X-ray structure of the Cj1427 in the presence of NADH and GDP-D-glycero-D-mannoheptose, an essential NAD-dependent dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative sugar-nucleotide epimerase/dehydratease, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S},5~{S},6~{S})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Spencer, K.D, Anderson, T.K, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
