4EE1
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to CTP and Manganese | | Descriptor: | BENZAMIDINE, CYTIDINE-5'-TRIPHOSPHATE, DNA primase, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4EDK
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to GTP and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4FM9
 
 | | Human topoisomerase II alpha bound to DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), DNA topoisomerase 2-alpha, ... | | Authors: | Wendorff, T.J, Schmidt, B.H, Heslop, P, Austin, C.A, Berger, J.M. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Structure of DNA-Bound Human Topoisomerase II Alpha: Conformational Mechanisms for Coordinating Inter-Subunit Interactions with DNA Cleavage.
J.Mol.Biol., 424, 2012
|
|
4EDG
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to ATP and Manganese | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BENZAMIDINE, DNA primase, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4E2K
 
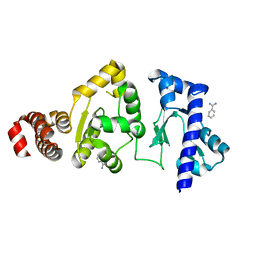 | | The structure of the S. aureus DnaG RNA Polymerase Domain | | Descriptor: | BENZAMIDINE, DNA primase | | Authors: | Rymer, R.U, Solorio, F.A, Clement, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-08 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4EDV
 
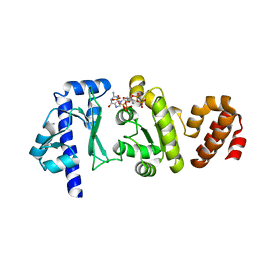 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to pppGpp and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4GFH
 
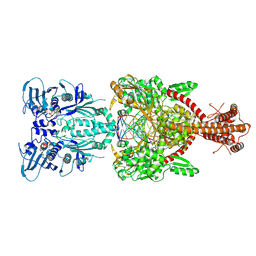 | | Topoisomerase II-DNA-AMPPNP complex | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*AP*CP*TP*GP*CP*TP*AP*C)-3'), ... | | Authors: | Schmidt, B.H, Osheroff, N, Berger, J.M. | | Deposit date: | 2012-08-03 | | Release date: | 2012-10-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (4.408 Å) | | Cite: | Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TO1
 
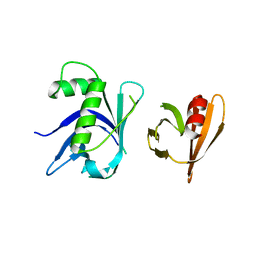 | | Two surfaces on Rtt106 mediate histone binding and chaperone activity | | Descriptor: | Histone chaperone RTT106 | | Authors: | Zunder, R.M, Antczak, A.J, Berger, J.M, Rine, J. | | Deposit date: | 2011-09-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UC1
 
 | | Mycobacterium tuberculosis gyrase type IIA topoisomerase C-terminal domain | | Descriptor: | ACETATE ION, CALCIUM ION, DNA gyrase subunit A, ... | | Authors: | Tretter, E.M, Berger, J.M. | | Deposit date: | 2011-10-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanisms for Defining Supercoiling Set Point of DNA Gyrase Orthologs: II. THE SHAPE OF THE GyrA SUBUNIT C-TERMINAL DOMAIN (CTD) IS NOT A SOLE DETERMINANT FOR CONTROLLING SUPERCOILING EFFICIENCY.
J.Biol.Chem., 287, 2012
|
|
4KM5
 
 | | X-ray crystal structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Berger, J.M, Doudna, J.A. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity.
Cell Rep, 3, 2013
|
|
3NUH
 
 | |
3R8F
 
 | | Replication initiator DnaA bound to AMPPCP and single-stranded DNA | | Descriptor: | 5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', Chromosomal replication initiator protein dnaA, MAGNESIUM ION, ... | | Authors: | Duderstadt, K.E, Chuang, K, Berger, J.M. | | Deposit date: | 2011-03-23 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.366 Å) | | Cite: | DNA stretching by bacterial initiators promotes replication origin opening.
Nature, 478, 2011
|
|
3RRN
 
 | | S. cerevisiae dbp5 l327v bound to gle1 h337r and ip6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
3RRM
 
 | | S. cerevisiae dbp5 l327v bound to nup159, gle1 h337r, ip6 and adp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
3PEU
 
 | | S. cerevisiae Dbp5 L327V C-terminal domain bound to Gle1 H337R and IP6 | | Descriptor: | ATP-dependent RNA helicase DBP5, GLYCEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEW
 
 | | S. cerevisiae Dbp5 L327V bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEV
 
 | | S. cerevisiae Dbp5 L327V C-terminal domain bound to Gle1 and IP6 | | Descriptor: | ATP-dependent RNA helicase DBP5, GLYCEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEY
 
 | | S. cerevisiae Dbp5 bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
4MEB
 
 | | Crystal structure of aCif-D158S | | Descriptor: | GLYCEROL, PHOSPHATE ION, Predicted protein | | Authors: | Bridges, A.A, Bahl, C.D, Madden, D.R. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Signature motifs identify an acinetobacter cif virulence factor with epoxide hydrolase activity.
J.Biol.Chem., 289, 2014
|
|
4DMF
 
 | |
4DMK
 
 | |
4DMH
 
 | |
6W7B
 
 | | K2P2.1 (TREK-1), 0 mM K+ | | Descriptor: | CADMIUM ION, DECANE, DODECANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
6W84
 
 | | K2P2.1 (TREK-1), 200 mM K+ | | Descriptor: | CADMIUM ION, DODECANE, N-OCTANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
6W8A
 
 | | K2P2.1 (TREK-1):ML335 complex, 10 mM K+ | | Descriptor: | CADMIUM ION, DODECANE, HEXADECANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
