7TKS
 
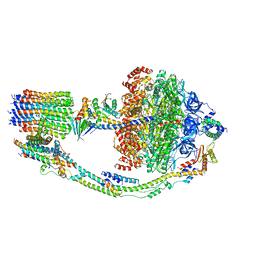 | |
7TKK
 
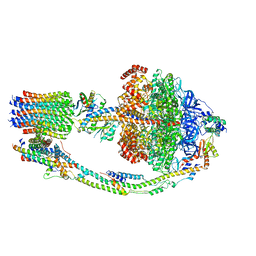 | |
7UWA
 
 | | Citrus V-ATPase State 1, H in contact with subunits AB | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Abdelaziz, R.A, Keon, K.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
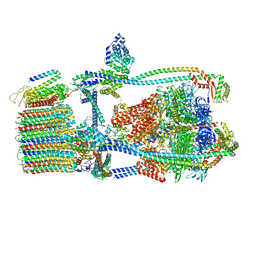
7UWC
 
 | | Citrus V-ATPase State 2, H in contact with subunit a | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
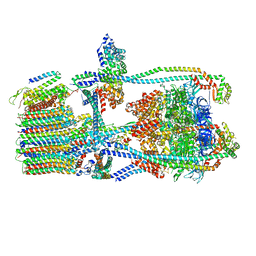
7UWD
 
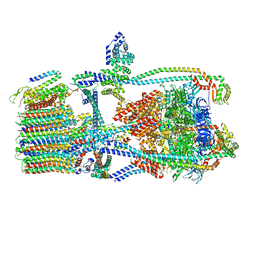 | | Citrus V-ATPase State 2, H in contact with subunits AB | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit, V-type proton ATPase subunit AP1 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UWB
 
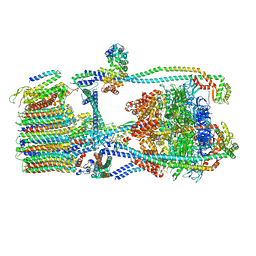 | | Citrus V-ATPase State 2, Highest-Resolution Class | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UW9
 
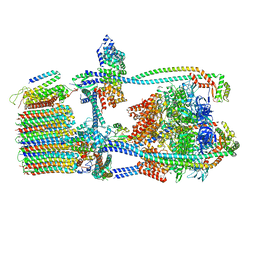 | | Citrus V-ATPase State 1, H in contact with subunit a | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7V05
 
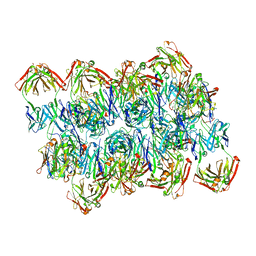 | | Complex of Plasmodium falciparum circumsporozoite protein with 850 Fab | | Descriptor: | 850 Fab Heavy Chain, 850 Fab Light Chain, Circumsporozoite protein | | Authors: | Kucharska, I, Prieto, K, Rubinstein, J.L, Julien, J.P. | | Deposit date: | 2022-05-09 | | Release date: | 2022-11-23 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-density binding to Plasmodium falciparum circumsporozoite protein repeats by inhibitory antibody elicited in mouse with human immunoglobulin repertoire.
Plos Pathog., 18, 2022
|
|
7KR2
 
 | |
6X87
 
 | | CryoEM structure of the Plasmodium berghei circumsporozoite protein in complex with inhibitory mouse antibody 3D11. | | Descriptor: | 3D11 Fab heavy chain, 3D11 Fab kappa chain, Circumsporozoite protein | | Authors: | Kucharska, I, Thai, E, Rubinstein, J, Julien, J.P. | | Deposit date: | 2020-06-01 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural ordering of the Plasmodium berghei circumsporozoite protein repeats by inhibitory antibody 3D11.
Elife, 9, 2020
|
|
6X8S
 
 | |
4EOZ
 
 | |
8D4T
 
 | | Mammalian CIV with GDN bound | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Di Trani, J, Rubinstein, J. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of mammalian complex IV inhibition by steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8U7Z
 
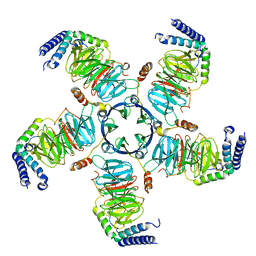 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(CTD)/Gbeta1gamma2 | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U83
 
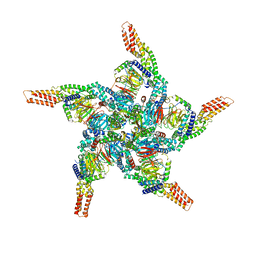 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.975 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U80
 
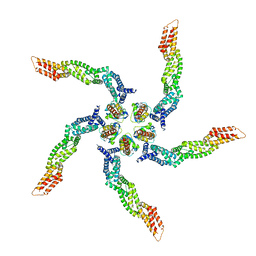 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(BTB)/Cullin3(NTD) | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U81
 
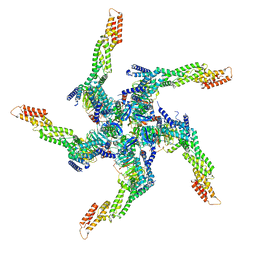 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U82
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State B From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6X8Q
 
 | |
6X8U
 
 | |
6X8P
 
 | |
2KX4
 
 | | Solution structure of Bacteriophage Lambda gpFII | | Descriptor: | Tail attachment protein | | Authors: | Maxwell, K.L, Cardarelli, L, Neudecker, P, Davidson, A.R, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-04-26 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Phages have adapted the same protein fold to fulfill multiple functions in virion assembly.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4K1O
 
 | |
4K1N
 
 | |
