8WZ3
 
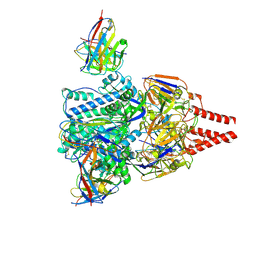 | | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 strain: A2) in complex with nAb 5B11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5B11 Fab Heavy Chain, 5B11 Fab Light Chain, ... | | Authors: | Liu, L, Sun, H, Sun, Y, Zheng, Q, Li, S, Zheng, Z, Xia, N. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-13 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | A potent broad-spectrum neutralizing antibody targeting a conserved region of the prefusion RSV F protein.
Nat Commun, 15, 2024
|
|
8WZ5
 
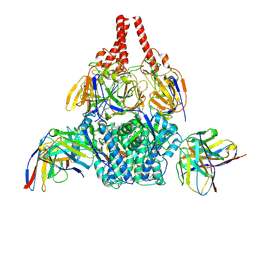 | | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 sc9-10 strain: B18537) in complex with humanized nAb 5B11 | | Descriptor: | 5B11 Fab Heavy Chain, 5B11 Fab Light Chain, RSV Fusion glycoprotein | | Authors: | Liu, L, Sun, H, Sun, Y, Zheng, Q, Li, S, Zheng, Z, Xia, N. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-13 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | A potent broad-spectrum neutralizing antibody targeting a conserved region of the prefusion RSV F protein.
Nat Commun, 15, 2024
|
|
8WZE
 
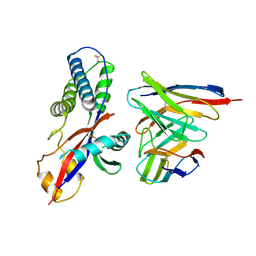 | | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 sc9-10 strain: B18537) in complex with humanized nAb 5B11 (localized refinement) | | Descriptor: | 5B11 Fab Heavy Chain, 5B11 Fab Light Chain, RSV pre-fusion glycoprotein | | Authors: | Liu, L, Sun, H, Sun, Y, Zheng, Q, Li, S, Zheng, Z, Xia, N. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A potent broad-spectrum neutralizing antibody targeting a conserved region of the prefusion RSV F protein.
Nat Commun, 15, 2024
|
|
8XSS
 
 | |
8XSR
 
 | |
8XST
 
 | |
3G11
 
 | | Structure of E. coli FabF(C163Q) in complex with dihydrophenyl platensimycin | | Descriptor: | 3-({3-[(1S,4S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxo-4-phenyldecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3G0Y
 
 | | Structure of E. coli FabF(C163Q) in complex with dihydroplatensimycin | | Descriptor: | 3-({3-[(1S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxodecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8ZNJ
 
 | | Cryo-EM structure of a short prokaryotic Argonaute system from archaeon Suldolobus islandicus | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*CP*AP*GP*GP*GP*TP*AP*TP*CP*TP*AP*AP*GP*CP*TP*TP*TP*GP*AP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Dai, Z.K, Guan, Z.Y, Han, W.Y, Zou, T.T. | | Deposit date: | 2024-05-27 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic insights into the activation of a short prokaryotic argonaute system from archaeon Sulfolobus islandicus.
Nucleic Acids Res., 53, 2025
|
|
5YY6
 
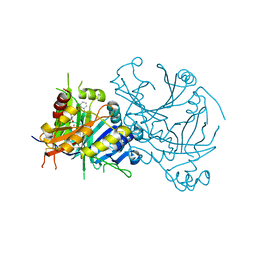 | | Crystal structure of Arabidopsis thaliana HPPD truncated mutant complexed with Benquitrione | | Descriptor: | 3-(2,6-dimethylphenyl)-1-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, W.C, Chen, J.N, Yang, G.F. | | Deposit date: | 2017-12-08 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal Structure of 4-Hydroxyphenylpyruvate Dioxygenase in Complex with Substrate Reveals a New Starting Point for Herbicide Discovery.
Research (Wash D C), 2019, 2019
|
|
3K1R
 
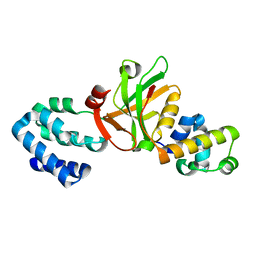 | |
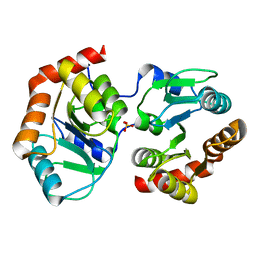
7YF5
 
 | |
2QP8
 
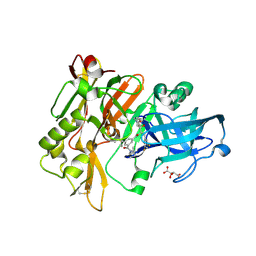 | | Structure of BACE Bound to SCH734723 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-[(2R,4S)-4-ETHOXYPIPERIDIN-2-YL]-2-HYDROXYETHYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C.O, Iserloh, U. | | Deposit date: | 2007-07-23 | | Release date: | 2008-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent pyrrolidine- and piperidine-based BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7WCL
 
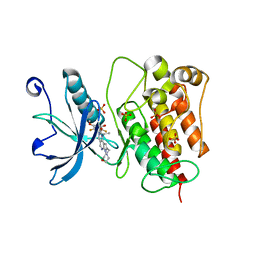 | | Crystal structure of FGFR1 kinase domain with Pemigatinib | | Descriptor: | 11-[2,6-bis(fluoranyl)-3,5-dimethoxy-phenyl]-13-ethyl-4-(morpholin-4-ylmethyl)-5,7,11,13-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),2(6),3,7-tetraen-12-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Lin, Q.M, Jiang, L.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Characterization of the cholangiocarcinoma drug pemigatinib against FGFR gatekeeper mutants.
Commun Chem, 5, 2022
|
|
6JIJ
 
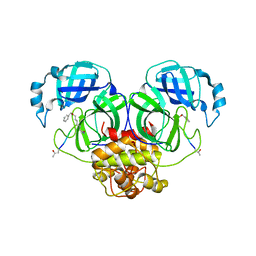 | |
2QK5
 
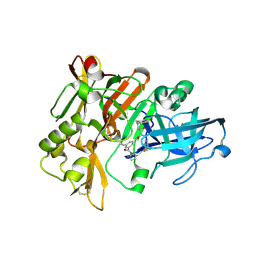 | | Structure of BACE1 bound to SCH626485 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C.O, Iserloh, U. | | Deposit date: | 2007-07-10 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent pyrrolidine- and piperidine-based BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5ZFZ
 
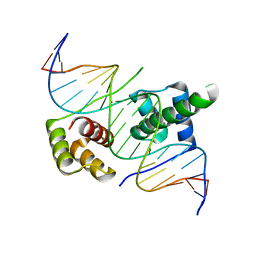 | | Crystal structure of human DUX4 homeodomains bound to A12T DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*TP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*AP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5YZ1
 
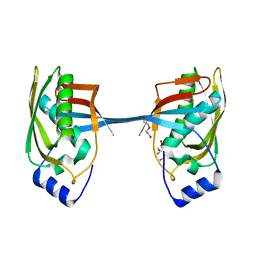 | | Crystal structure of human Archease | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein archease | | Authors: | Duan, S.Y, Li, J.X. | | Deposit date: | 2017-12-11 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of human archease, a key cofactor of tRNA splicing ligase complex.
Int.J.Biochem.Cell Biol., 122, 2020
|
|
5ZYT
 
 | |
2QMF
 
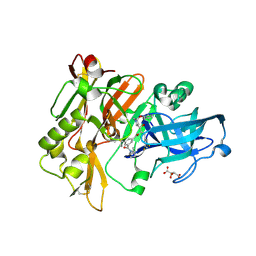 | | Structure of BACE Bound to SCH735310 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-2-[(2R,4R)-4-PHENOXYPYRROLIDIN-2-YL]ETHYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C.O, Iserloh, U. | | Deposit date: | 2007-07-16 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent pyrrolidine- and piperidine-based BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2QVR
 
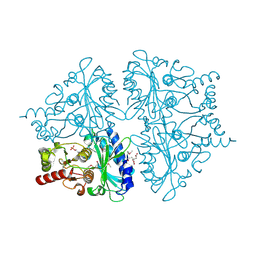 | | E. coli Fructose-1,6-bisphosphatase: Citrate, Fru-2,6-P2, and Mg2+ bound | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CITRIC ACID, Fructose-1,6-bisphosphatase, ... | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of Mammalian and Bacterial Fructose-1,6-bisphosphatase Reveal the Basis for Synergism in AMP/Fructose 2,6-Bisphosphate Inhibition.
J.Biol.Chem., 282, 2007
|
|
5ZYU
 
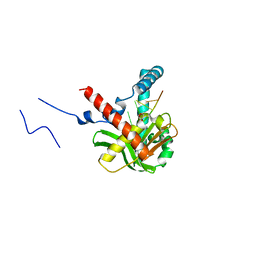 | | The crystal structure of humanMGME1 with single strand DNA2 | | Descriptor: | DNA (5'-D(P*CP*AP*AP*CP*AP*AP*CP*A)-3'), GLYCEROL, Mitochondrial genome maintenance exonuclease 1 | | Authors: | Yang, C, Gan, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural insights into DNA degradation by human mitochondrial nuclease MGME1
Nucleic Acids Res., 46, 2018
|
|
2F4B
 
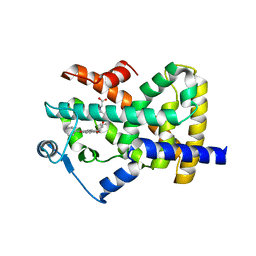 | | Crystal structure of the ligand binding domain of human PPAR-gamma in complex with an agonist | | Descriptor: | (5-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPOXY}-1H-INDOL-1-YL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Lu, I.L, Peng, Y.H, Mahindroo, N, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2005-11-23 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Indol-1-yl Acetic Acids as Peroxisome Proliferator-Activated Receptor Agonists: Design, Synthesis, Structural Biology, and Molecular Docking Studies
J.Med.Chem., 49, 2006
|
|
5ZYW
 
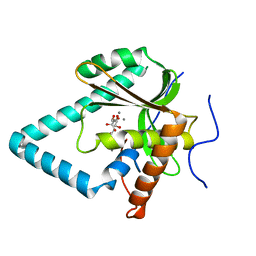 | | The crystal structure of apo-HsMGME1 with Mn2+ | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Yang, C, Gan, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into DNA degradation by human mitochondrial nuclease MGME1
Nucleic Acids Res., 46, 2018
|
|
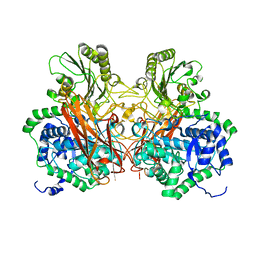
5Z9S
 
 | |
