7LUY
 
 | |
4QIC
 
 | |
8S9G
 
 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
4NPS
 
 | | Crystal Structure of Bep1 protein (VirB-translocated Bartonella effector protein) from Bartonella clarridgeiae | | Descriptor: | ACETATE ION, Bartonella effector protein (Bep) substrate of VirB T4SS | | Authors: | Dranow, D.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolutionary Diversification of Host-Targeted Bartonella Effectors Proteins Derived from a Conserved FicTA Toxin-Antitoxin Module.
Microorganisms, 9, 2021
|
|
9DNE
 
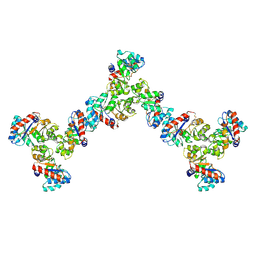 | | Pseudosymmetric protein nanocage GI9-F7 (local refinement) | | Descriptor: | Pseudosymmetric protein nanocage GI9-F7 A chain, Pseudosymmetric protein nanocage GI9-F7 B chain, Pseudosymmetric protein nanocage GI9-F7 C chain | | Authors: | Park, Y.J, Dowling, Q.M, King, N.P, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-09-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Hierarchical design of pseudosymmetric protein nanocages.
Nature, 638, 2025
|
|
4NOZ
 
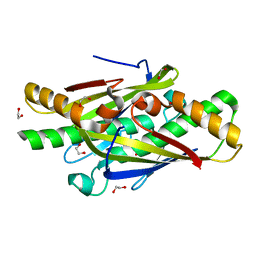 | | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Organic hydroperoxide resistance protein | | Authors: | Dranow, D.M, Lukacs, C.M, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia
TO BE PUBLISHED
|
|
9C45
 
 | |
9C44
 
 | |
8TVI
 
 | |
8TVB
 
 | |
8TVF
 
 | |
8TVE
 
 | |
8TVG
 
 | |
8TVH
 
 | | Langya henipavirus postfusion F protein in complex with 4G5 Fab, local refinement of the viral membrane proximal region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4G5 heavy chain, 4G5 light chain, ... | | Authors: | Wang, Z, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-08-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and design of Langya virus glycoprotein antigens.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3IEW
 
 | |
9DND
 
 | | Pseudosymmetric protein nanocage GI4 -F7 (local refinement) | | Descriptor: | Pseudosymmetric protein nanocages GI4 A Chain, Pseudosymmetric protein nanocages GI4 B Chain, Pseudosymmetric protein nanocages GI4 C Chain | | Authors: | Park, Y.J, Dowling, Q.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), King, N.P, Veesler, D. | | Deposit date: | 2024-09-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Hierarchical design of pseudosymmetric protein nanocages.
Nature, 638, 2025
|
|
8UR5
 
 | |
3U0F
 
 | |
8UR7
 
 | |
7KQ6
 
 | |
6BFU
 
 | | Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein, ... | | Authors: | Xiong, X, Tortorici, M.A, Snijder, S, Yoshioka, C, Walls, A.C, Li, W, Seattle Structural Genomics Center for Infectious Disease (SSGCID), McGuire, A.T, Rey, F.A, Bosch, B.J, Veesler, D. | | Deposit date: | 2017-10-27 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Glycan Shield and Fusion Activation of a Deltacoronavirus Spike Glycoprotein Fine-Tuned for Enteric Infections.
J. Virol., 92, 2018
|
|
7LBJ
 
 | |
7L6P
 
 | |
7LTQ
 
 | |
3LRF
 
 | |
