7DNH
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 2H3 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of 2H3 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNK
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 5G9 | | Descriptor: | Major capsid protein L1, The heavy chain of 5G9 Fab fragment, The light chain of 5G9 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (6.41 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNL
 
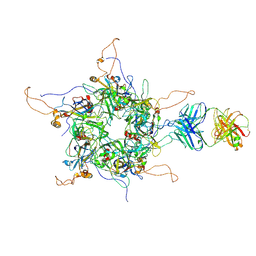 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of A4B4 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of A4B4 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DBF
 
 | |
7DC9
 
 | |
7DCA
 
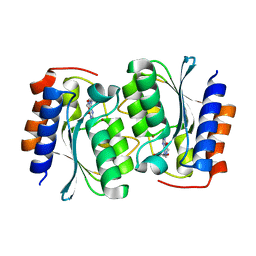 | | The structure of the Arabidopsis thaliana guanosine deaminase bound by xanthosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-10-23 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The C-terminal loop of Arabidopsis thaliana guanosine deaminase is essential to catalysis.
Chem.Commun.(Camb.), 57, 2021
|
|
7DGY
 
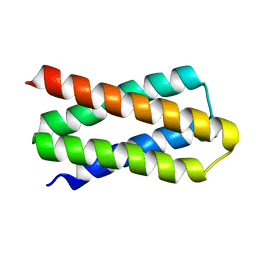 | | De novo designed protein H4C2R | | Descriptor: | de novo designed protein H4C2R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DH1
 
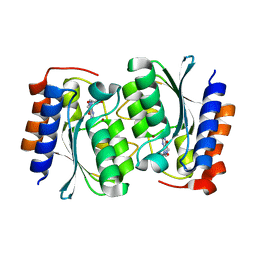 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N2-Methylguanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-12 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DQN
 
 | |
7DM6
 
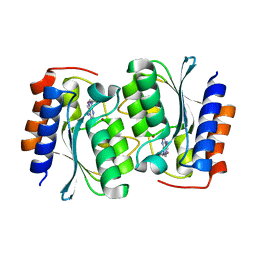 | | The structure of the Arabidopsis thaliana guanosine deaminase complexed with crotonoside | | Descriptor: | 6-azanyl-9-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1H-purin-2-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DM5
 
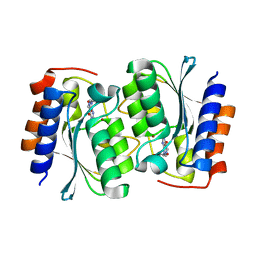 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with guanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DGC
 
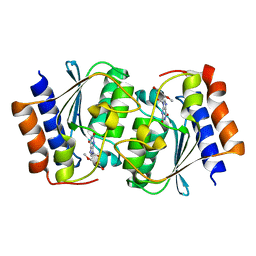 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with 2'-O-Methylguanosine | | Descriptor: | 9-[(2R,3R,4R)-5-(hydroxymethyl)-3-methoxy-4-oxidanyl-oxolan-2-yl]-3H-purine-2,6-dione, Guanosine deaminase, SODIUM ION, ... | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7EO0
 
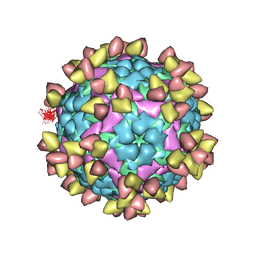 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
7U87
 
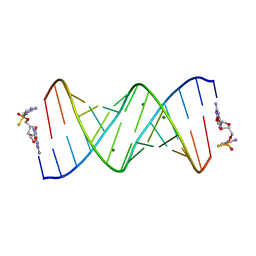 | | Product of 13mer primer with activated G monomer diastereomer 1 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*(G46)P*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
7U88
 
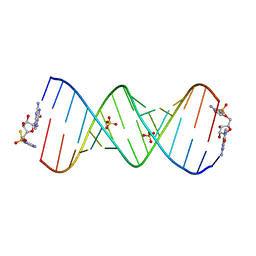 | | Product of 13mer primer with activated G monomer diastereomer 2 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*(G46)P*(G46))-3'), SULFATE ION | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
7U89
 
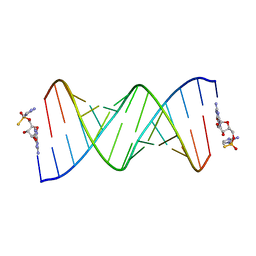 | | Product of 14mer primer with activated G monomer diastereomer 1 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
7U8A
 
 | | Product of 14mer primer with activated G monomer diastereomer 2 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
7UHE
 
 | |
7WIE
 
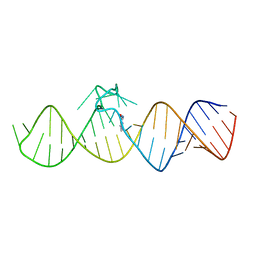 | | The THF-II riboswitch bound to 7DG | | Descriptor: | 7-DEAZAGUANINE, RNA (50-MER) | | Authors: | Xu, L, Fang, X, Xiao, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WII
 
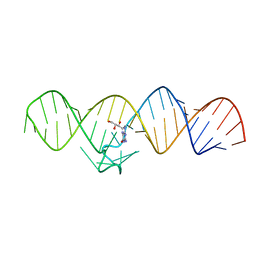 | | The THF-II riboswitch bound to NPR | | Descriptor: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, RNA (50-MER) | | Authors: | Xu, L, Fang, X, Xiao, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WIA
 
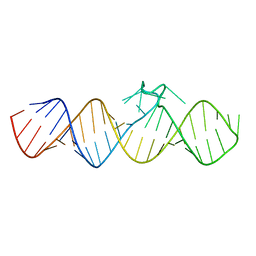 | | The apo-form of THF-II C22G riboswitch | | Descriptor: | RNA (50-MER) | | Authors: | Xu, L, Fang, X, Xiao, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WIB
 
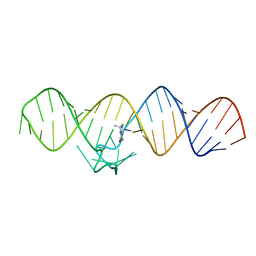 | | The THF-II riboswitch bound to THF | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, RNA (50-MER) | | Authors: | Xu, L, Fang, X, Xiao, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WI9
 
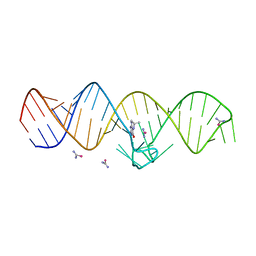 | | The THF-II riboswitch bound to THF and soaking with SeUrea | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, RNA (50-MER), selenourea | | Authors: | Xu, L, Fang, X, Xiao, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7XDI
 
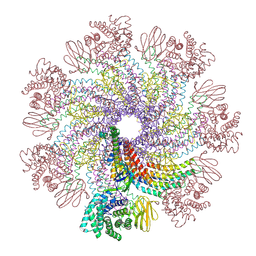 | | Tail structure of bacteriophage SSV19 | | Descriptor: | B210, C131, VP1, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2022-03-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into a spindle-shaped archaeal virus with a sevenfold symmetrical tail.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WIF
 
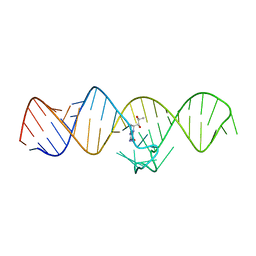 | | The THF-II riboswitch bound to H4B | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, RNA (50-MER) | | Authors: | Xu, L, Fang, X, Xiao, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
