7SYC
 
 | |
4Q4L
 
 | |
6UWW
 
 | |
6W15
 
 | |
4OO0
 
 | |
6X79
 
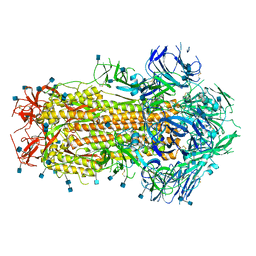 | | Prefusion SARS-CoV-2 S ectodomain trimer covalently stabilized in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | McCallum, M, Walls, A.C, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-05-29 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W14
 
 | |
7SZS
 
 | |
7SZV
 
 | |
5DB4
 
 | |
5DLC
 
 | |
4PQ9
 
 | |
5UCR
 
 | |
5UMF
 
 | |
6X9F
 
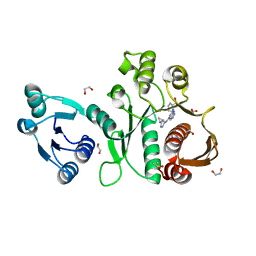 | | Pseudomonas aeruginosa MurC with AZ8074 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ8074
To Be Published
|
|
4FRY
 
 | |
7MYQ
 
 | |
7MYS
 
 | |
8ERR
 
 | |
8ERQ
 
 | |
6NXG
 
 | | Crystal structure of glycylpeptide N-tetradecanoyltransferase from Plasmodium vivax in complex with inhibitor 303a | | Descriptor: | 1,2-ETHANEDIOL, 5-(4-chlorophenyl)-3-({[3-(morpholine-4-carbonyl)phenyl]amino}methyl)pyridin-2(1H)-one, BETA-MERCAPTOETHANOL, ... | | Authors: | Staker, B.L, Mayclin, S, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Selective Inhibitors ofPlasmodiumN-Myristoyltransferase by High-Throughput Screening.
J.Med.Chem., 63, 2020
|
|
6O5D
 
 | | PYOCHELIN | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, SULFATE ION | | Authors: | Rupert, P.B, Strong, R.K, Clifton, M.C, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
4EFZ
 
 | |
8VGT
 
 | |
8VKA
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 9c | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
