3EUL
 
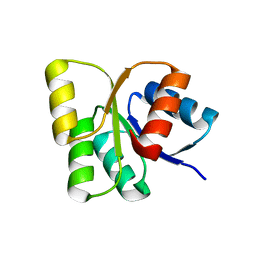 | | Structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, POSSIBLE NITRATE/NITRITE RESPONSE TRANSCRIPTIONAL REGULATORY PROTEIN NARL (DNA-binding response regulator, LuxR family) | | Authors: | Schneider, G, Schnell, R, Agren, D. | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4NP8
 
 | | Structure of an amyloid forming peptide VQIVYK from the second repeat region of tau (alternate polymorph) | | Descriptor: | Microtubule-associated protein tau | | Authors: | Landau, M, Eisenberg, D, Sawaya, M.R, Dannenberg, J, Kobko, N. | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular mechanisms for protein-encoded inheritance.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4B5B
 
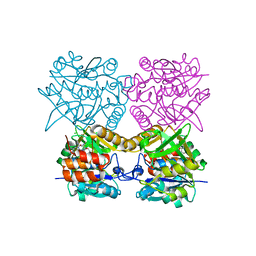 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, N-(6-amino-1-benzyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-N,3-dimethylbenzenesulfonamide | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-08-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
3KVI
 
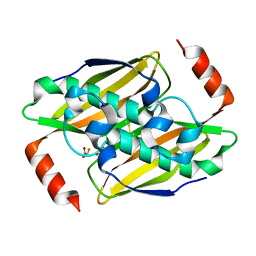 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - T42A mutant in complex with fluoro-acetate | | Descriptor: | Fluoroacetyl-CoA thioesterase FlK, fluoroacetic acid | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
4AUE
 
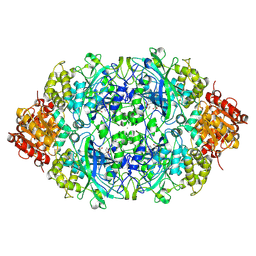 | | Crystal structure, recombinant expression and mutagenesis studies of the bifunctional catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yuzugullu, Y, Trinh, C.H, Smith, M.A, Pearson, A.R, Phillips, S.E.V, Sutay Kocabas, D, Bakir, U, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-05-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure, Recombinant Expression and Mutagenesis Studies of the Catalase with Oxidase Activity from Scytalidium Thermophilum
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4AUP
 
 | | Tuber borchii Phospholipase A2 | | Descriptor: | ACETATE ION, PHOSPHOLIPASE A2 GROUP XIII, THIOCYANATE ION | | Authors: | Cavazzini, D, Meschi, F, Corsini, R, Bolchi, A, Rossi, G.-L, Einsle, O, Ottonello, S. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-12 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoproteolytic Activation of a Symbiosis-Regulated Truffle Phospholipase A2
J.Biol.Chem., 288, 2013
|
|
3KVZ
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thiesterase FlK - wild type FlK in complex with FAcCPan | | Descriptor: | (2R)-N-{3-[(5-fluoro-4-oxopentyl)amino]-3-oxopropyl}-2,4-dihydroxy-3,3-dimethylbutanamide, Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
1HCE
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
4B0S
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEAMIDASE-DEPUPYLASE DOP, MAGNESIUM ION | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
3KZH
 
 | |
2PCS
 
 | | Crystal structure of conserved protein from Geobacillus kaustophilus | | Descriptor: | Conserved protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Wu, B, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of conserved protein from Geobacillus kaustophilus
To be Published
|
|
3L04
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase E92P mutant complexed with carbamyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
3L0L
 
 | |
3EWY
 
 | | K314A mutant of human orotidyl-5'-monophosphate decarboxylase soaked with OMP, decarboxylated to UMP | | Descriptor: | GLYCEROL, Orotidine-5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Heinrich, D, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
4BAM
 
 | | Thrombin in complex with inhibitor | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-1-[(2R)-2-cyclohexyl-2-[[2-(dimethylamino)-2-oxidanylidene-ethyl]amino]ethanoyl]azetidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-1, ... | | Authors: | Xue, Y, Musil, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification of Structure-Kinetic and Structure-Thermodynamic Relationships for Thrombin Inhibitors.
Biochemistry, 52, 2013
|
|
2OL9
 
 | |
4BAH
 
 | | Thrombin in complex with inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-1, SODIUM ION, ... | | Authors: | Xue, Y, Musil, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of Structure-Kinetic and Structure-Thermodynamic Relationships for Thrombin Inhibitors.
Biochemistry, 52, 2013
|
|
2OMP
 
 | |
4B94
 
 | | Crystal structure of human Mps1 TPR domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, DUAL SPECIFICITY PROTEIN KINASE TTK, GLYCEROL, ... | | Authors: | Littler, D, von Castelmur, E, De Marco, V, Perrakis, A. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Tpr Domain-Containing N-Terminal Module of Mps1 is Required for its Kinetochore Localization by Aurora B.
J.Cell Biol., 201, 2013
|
|
3L4S
 
 | | Crystal structure of C151G mutant of Glyceraldehyde 3-phosphate dehydrogenase 1 (GAPDH1) from methicillin resistant Staphylococcus aureus MRSA252 complexed with NAD and G3P | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-12-21 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3EX1
 
 | | human orotidyl-5'-monophosphate decarboxylase soaked with 6-cyano-UMP, converted to UMP | | Descriptor: | 6-cyanouridine 5'-phosphate, GLYCEROL, Orotidine-5'-phosphate decarboxylase, ... | | Authors: | Heinrich, D, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
4BAQ
 
 | | Thrombin in complex with inhibitor | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-1-[(2R)-2-cyclohexyl-2-[[2-(ethylamino)-2-oxidanylidene-ethyl]amino]ethanoyl]azetidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-1, ... | | Authors: | Xue, Y, Musil, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Identification of Structure-Kinetic and Structure-Thermodynamic Relationships for Thrombin Inhibitors.
Biochemistry, 52, 2013
|
|
4B0O
 
 | | Crystal structure of soman-aged human butyrylcholinesterase in complex with benzyl pyridinium-4-methyltrichloroacetimidate | | Descriptor: | 1-BENZYL-4-({[(1E)-2,2,2-TRICHLOROETHANIMIDOYL]OXY}METHYL)PYRIDINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wandhammer, M, de Koning, M, van Grol, M, Noort, D, Goeldner, M, Nachon, F. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Step Toward the Reactivation of Aged Cholinesterases -Crystal Structure of Ligands Binding to Aged Human Butyrylcholinesterase
Chem.Biol.Interact, 203, 2013
|
|
1GUI
 
 | | CBM4 structure and function | | Descriptor: | CALCIUM ION, GLYCEROL, LAMINARINASE 16A, ... | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-27 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1H64
 
 | |
