3I1C
 
 | |
3HYJ
 
 | | Crystal structure of the N-terminal LAGLIDADG domain of DUF199/WhiA | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DUF199/WhiA, ... | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease.
Structure, 17, 2009
|
|
3HYI
 
 | | Crystal structure of full-length DUF199/WhiA from Thermatoga maritima | | Descriptor: | GLYCEROL, Protein DUF199/WhiA, SODIUM ION | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease
Structure, 17, 2009
|
|
3KO2
 
 | | I-MsoI re-designed for altered DNA cleavage specificity (-7C) | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*GP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*CP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2009-11-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
3LDY
 
 | | An extraordinary mechanism of DNA perturbation exhibited by the rare-cutting HNH restriction endonuclease PacI | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*A)-3'), DNA (5'-D(P*AP*TP*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Heiter, D, Chan, S.-H, Xu, S.-Y, Wilson, G, Stoddard, B.L. | | Deposit date: | 2010-01-13 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
3S1S
 
 | | Characterization and crystal structure of the type IIG restriction endonuclease BpuSI | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Shen, B.W, Xu, D, Chan, S.-H, Zheng, Y, Zhu, Y, Xu, S.-Y, Stoddard, B.L. | | Deposit date: | 2011-05-16 | | Release date: | 2011-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI.
Nucleic Acids Res., 39, 2011
|
|
3MIS
 
 | | I-MsoI re-designed for altered DNA cleavage specificity (-8G) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
3M7K
 
 | | Crystal structure of PacI-DNA Enzyme product complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*AP*AP*T)-3'), DNA (5'-D(P*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
3MIP
 
 | | I-MsoI re-designed for altered DNA cleavage specificity (-8GCG) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*CP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2010-04-11 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
3UGM
 
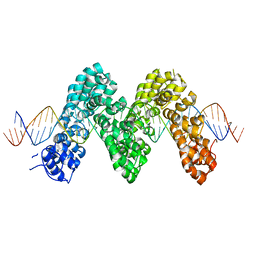 | | Structure of TAL effector PthXo1 bound to its DNA target | | Descriptor: | DNA-1, DNA-2, TAL effector AvrBs3/PthA | | Authors: | Mak, A.N.S, Bradley, P, Cernadas, R.A, Bogdanove, A.J, Stoddard, B.L. | | Deposit date: | 2011-11-02 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of TAL Effector PthXo1 Bound to Its DNA Target.
Science, 335, 2012
|
|
3MX1
 
 | |
3MX4
 
 | | DNA binding and cleavage by the GIY-YIG endonuclease R.Eco29KI inactive variant E142Q | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*AP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*GP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*CP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*TP*CP*CP*CP*G)-3'), Eco29kIR | | Authors: | Mak, A.N.S, Lambert, A.R, Stoddard, B.L. | | Deposit date: | 2010-05-06 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Folding, DNA Recognition, and Function of GIY-YIG Endonucleases: Crystal Structures of R.Eco29kI.
Structure, 18, 2010
|
|
3R3P
 
 | |
3T1G
 
 | |
4YIT
 
 | |
4YIS
 
 | | Crystal Structure of LAGLIDADG Meganuclease I-CpaMI Bound to Uncleaved DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (28-MER), ... | | Authors: | Hallinan, J.P, Kaiser, B.K, Stoddard, B.L. | | Deposit date: | 2015-03-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity.
Structure, 24, 2016
|
|
4YHX
 
 | |
4Z1Z
 
 | |
4YXX
 
 | |
4YXY
 
 | |
4Z20
 
 | |
4YY5
 
 | |
4YY2
 
 | |
4YXZ
 
 | |
4R58
 
 | |
