2Z2F
 
 | | X-ray Crystal Structure of Bovine Stomach Lysozyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme C-2, SODIUM ION | | Authors: | Akieda, D, Nonaka, Y, Watanabe, N, Tanaka, I, Kamiya, M, Aizawa, T, Nitta, K, Demura, M, Kawano, K. | | Deposit date: | 2007-05-21 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stability of Bovine Stomach Lysozyme in Acidic Condition
To be Published
|
|
2ZOY
 
 | | The multi-drug binding transcriptional repressor CgmR (CGL2612 protein) from C.glutamicum | | Descriptor: | GLYCEROL, Transcriptional regulator | | Authors: | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The CGL2612 protein from Corynebacterium glutamicum is a drug resistance-related transcriptional repressor: structural and functional analysis of a newly identified transcription factor from genomic DNA analysis
J.Biol.Chem., 280, 2005
|
|
2ZCX
 
 | |
3A3C
 
 | | Crystal structure of TIM40/MIA40 fusing MBP, C296S and C298S mutant | | Descriptor: | Maltose-binding periplasmic protein, LINKER, Mitochondrial intermembrane space import and assembly protein 40, ... | | Authors: | Kawano, S, Naoe, M, Momose, T, Watanabe, N, Endo, T. | | Deposit date: | 2009-06-11 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZXT
 
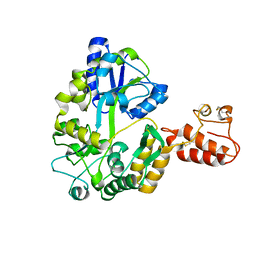 | | Crystal structure of Tim40/MIA40, a disulfide relay system in mitochondria, solved as MBP fusion protein | | Descriptor: | Maltose-binding periplasmic protein, LINKER, Mitochondrial intermembrane space import and assembly protein 40, ... | | Authors: | Kawano, S, Momose, T, Watanabe, N, Endo, T. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZOS
 
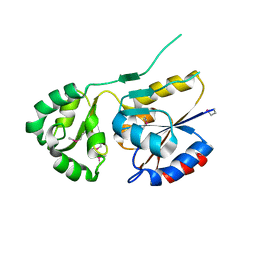 | |
2ZU7
 
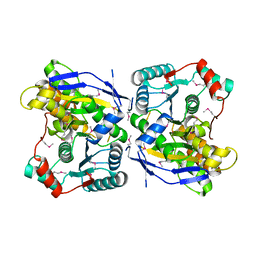 | |
2YZY
 
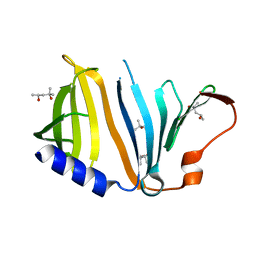 | | Crystal structure of uncharacterized conserved protein from Thermus thermophilus HB8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Putative uncharacterized protein TTHA1012 | | Authors: | Ebihara, A, Watanabe, N, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Thermus thermophilus HB8
To be Published
|
|
2ZB9
 
 | | Crystal structure of TetR family transcription regulator SCO0332 | | Descriptor: | Putative transcriptional regulator | | Authors: | Okada, U, Kondo, K, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional analysis of the TetR-family transcriptional regulator SCO0332 from Streptomyces coelicolor
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
2ZU9
 
 | |
2ZU8
 
 | |
2DFC
 
 | | Xylanase II from Tricoderma reesei at 293K | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Harata, K, Akiba, T. | | Deposit date: | 2006-02-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DFB
 
 | | Xylanase II from Tricoderma reesei at 100K | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, SULFATE ION | | Authors: | Harata, K, Akiba, T. | | Deposit date: | 2006-02-28 | | Release date: | 2006-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3QUG
 
 | | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Gallium-porphyrin | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING GA, ... | | Authors: | Moriwaki, Y, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-02-24 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of recognition of antibacterial porphyrins by heme-transporter IsdH-NEAT3 of Staphylococcus aureus.
Biochemistry, 50, 2011
|
|
3QUH
 
 | |
3M4Y
 
 | | Structural characterization of the subunit A mutant P235A of the A-ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Manimekalai, M.S, Balakrishna, A.M, Kumar, A, Priya, R, Biukovic, G, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-03-12 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
1J0A
 
 | | Crystal Structure Analysis of the ACC deaminase homologue | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, ISOPROPYL ALCOHOL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fujino, A, Ose, T, Honma, M, Yao, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and enzymatic properties of 1-aminocyclopropane-1-carboxylate deaminase homologue from Pyrococcus horikoshii
J.Mol.Biol., 341, 2004
|
|
1J0E
 
 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
3MFY
 
 | | Structural characterization of the subunit A mutant F236A of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Balakrishna, A.M, Kumar, A, Manimekali, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-04-05 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
7TVW
 
 | | Crystal structure of Arabidopsis thaliana DLK2 | | Descriptor: | Alpha/beta-Hydrolases superfamily protein | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Arabidopsis DWARF14-LIKE2 (DLK2) reveals a distinct substrate binding pocket architecture.
Plant Direct, 6, 2022
|
|
1J0C
 
 | | ACC deaminase mutated to catalytic residue | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1J0D
 
 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1J0B
 
 | | Crystal Structure Analysis of the ACC deaminase homologue complexed with inhibitor | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Fujino, A, Ose, T, Honma, M, Yao, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and enzymatic properties of 1-aminocyclopropane-1-carboxylate deaminase homologue from Pyrococcus horikoshii
J.Mol.Biol., 341, 2004
|
|
2E7D
 
 | | Crystal structure of a NEAT domain from Staphylococcus aureus | | Descriptor: | ACETATE ION, GLYCEROL, Hypothetical protein IsdH, ... | | Authors: | Suenaga, A, Tanaka, Y, Yao, M, Kumagai, I, Tanaka, I, Tsumoto, K. | | Deposit date: | 2007-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for multimeric heme complexation through a specific protein-heme interaction: the case of the third neat domain of IsdH from Staphylococcus aureus
J.Biol.Chem., 283, 2008
|
|
2RD3
 
 | |
