8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.03 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC2
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 1 YB9-258 Fab (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8E8I
 
 | |
8EC5
 
 | |
7Y99
 
 | | Crystal Structure Analysis of cp2 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, CP2 peptide, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YAA
 
 | | Crystal structure analysis of cp3 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, GLYCEROL, N-(2-acetamidoethyl)-4-(4-methanoyl-1,3-thiazol-2-yl)benzamide, ... | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y90
 
 | | Crystal Structure Analysis of cp1 bound BCL2 | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W. | | Deposit date: | 2022-06-24 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YA5
 
 | | Crystal structure analysis of cp1 bound BCL2/G101V | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YB7
 
 | | anti-apoptotic protein BCL-2-M12 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide, cp2 peptide | | Authors: | Li, F.W, Liu, C, Wu, D.L. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y8D
 
 | | Crystal structure of cp1 bound BCLxl | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Bcl-2-like protein 1, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
5W2E
 
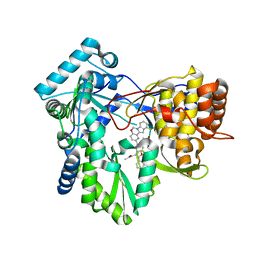 | | HCV NS5B RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor MK-8876 | | Descriptor: | 2-(4-fluorophenyl)-5-(11-fluoro-6H-pyrido[2',3':5,6][1,3]oxazino[3,4-a]indol-2-yl)-N-methyl-6-[methyl(methylsulfonyl)amino]-1-benzofuran-3-carboxamide, Genome polyprotein | | Authors: | Lesburg, C.A, Ummat, A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a New Structural Class of Broadly Acting HCV Non-Nucleoside Inhibitors Leading to the Discovery of MK-8876.
ChemMedChem, 12, 2017
|
|
8KEB
 
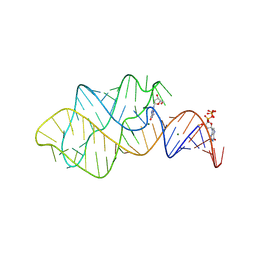 | | Crystal structure of 2'-dG-III riboswitch with 2'-dG | | Descriptor: | 2'-DEOXY-GUANOSINE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liao, W, Huang, L. | | Deposit date: | 2023-08-11 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A general strategy for engineering GU base pairs to facilitate RNA crystallization.
Nucleic Acids Res., 53, 2025
|
|
6AH3
 
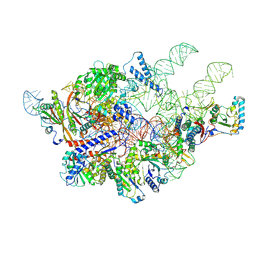 | | Cryo-EM structure of yeast Ribonuclease P with pre-tRNA substrate | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
6N6O
 
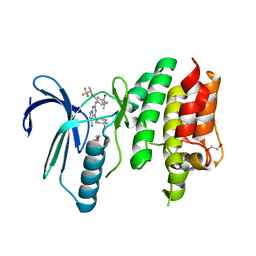 | | Crystal structure of the human TTK in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({5-chloro-4-[(cis-4-hydroxy-4-methylcyclohexyl)oxy]-7H-pyrrolo[2,3-d]pyrimidin-2-yl}amino)-N,N-dimethyl-3-{[(2R)-1,1,1-trifluoropropan-2-yl]oxy}benzamide, Dual specificity protein kinase TTK, ... | | Authors: | Fenalti, G. | | Deposit date: | 2018-11-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design and Optimization Leading to an Orally Active TTK Protein Kinase Inhibitor with Robust Single Agent Efficacy.
J.Med.Chem., 62, 2019
|
|
8ZHQ
 
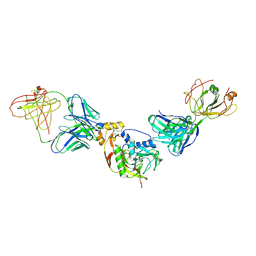 | | SFTSV Gn in complex with JK-8/12 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, ... | | Authors: | Shang, H, Guo, Y, Zhang, N, Liu, W, Li, H. | | Deposit date: | 2024-05-11 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Discovery and characterization of potent broadly neutralizing antibodies from human survivors of severe fever with thrombocytopenia syndrome.
Ebiomedicine, 111, 2024
|
|
6AGB
 
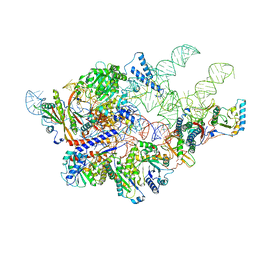 | | Cryo-EM structure of yeast Ribonuclease P | | Descriptor: | RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, Ribonuclease P protein subunit RPR2, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
8TWX
 
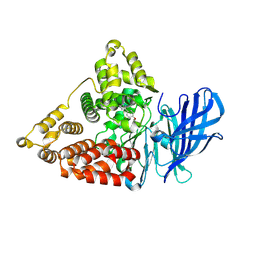 | | Synthesis and Evaluation of Diaryl Ether Modulators of the Leukotriene A4 Hydrolase Aminopeptidase Activity | | Descriptor: | 5-[4-(4-chlorophenoxy)phenyl]-1H-pyrazol-3-amine, Leukotriene A-4 hydrolase, ZINC ION | | Authors: | Lee, K.H, Lee, S.H, Paige, M, Noble, S.M. | | Deposit date: | 2023-08-21 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis and Evaluation of diaryl ether modulators of the leukotriene A 4 hydrolase aminopeptidase activity.
Eur.J.Med.Chem., 272, 2024
|
|
8Z9K
 
 | | Crystal structure of LINE-1 ribozyme | | Descriptor: | RNA (41-MER), SODIUM ION | | Authors: | Ren, Y, Huang, L, Lin, X. | | Deposit date: | 2024-04-23 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.86 Å) | | Cite: | A general strategy for engineering GU base pairs to facilitate RNA crystallization.
Nucleic Acids Res., 53, 2025
|
|
8Z8U
 
 | |
8Z8Q
 
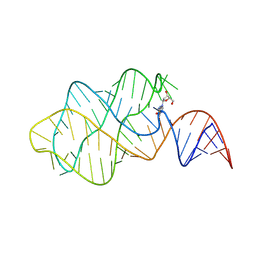 | | Crystal structure of 2'-dG riboswitch | | Descriptor: | 2'-DEOXY-GUANOSINE, RNA (71-MER) | | Authors: | Liao, W, Ren, Y, Huang, L. | | Deposit date: | 2024-04-22 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A general strategy for engineering GU base pairs to facilitate RNA crystallization.
Nucleic Acids Res., 53, 2025
|
|
8ZA0
 
 | | Crystal structure of OR4K15 ribozyme | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*UP*GP*GP*GP*CP*CP*AP*GP*GP*CP*AP*GP*G)-3') | | Authors: | Ren, Y, Huang, L. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A general strategy for engineering GU base pairs to facilitate RNA crystallization.
Nucleic Acids Res., 53, 2025
|
|
8ZAU
 
 | |
8ZA4
 
 | | Crystal structure of OR4K15 ribozyme mutant - chainBDF C2U | | Descriptor: | RNA (5'-R(*CP*UP*UP*GP*CP*GP*CP*UP*CP*AP*AP*CP*CP*AP*GP*A)-3'), RNA (5'-R(*UP*CP*UP*GP*GP*GP*CP*CP*AP*GP*GP*CP*AP*GP*G)-3') | | Authors: | Ren, Y, Huang, L. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A general strategy for engineering GU base pairs to facilitate RNA crystallization.
Nucleic Acids Res., 53, 2025
|
|
8XJV
 
 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
9KPH
 
 | |
