9JTA
 
 | |
9JW1
 
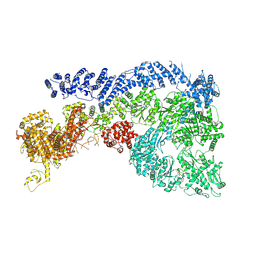 | | Cryo-EM structure of Human RNF213 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ring finger protein 213 | | Authors: | Zhang, H. | | Deposit date: | 2024-10-09 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Shigella effector IpaH1.4 subverts host E3 ligase RNF213 to evade antibacterial immunity.
Nat Commun, 16, 2025
|
|
1SZJ
 
 | |
4WXM
 
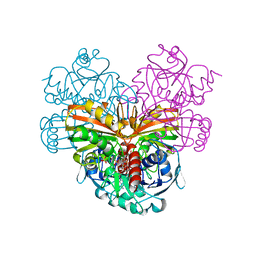
 | | FleQ REC domain from Pseudomonas aeruginosa PAO1 | | Descriptor: | Transcriptional regulator FleQ | | Authors: | Su, T, Liu, S, Gu, L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The REC domain mediated dimerization is critical for FleQ from Pseudomonas aeruginosa to function as a c-di-GMP receptor and flagella gene regulator
J.Struct.Biol., 192, 2015
|
|
9BCE
 
 | | Shewanella oneidensis LysR family regulator SO0839 regulatory domain | | Descriptor: | Transcriptional regulator LysR family | | Authors: | Han, S.R, Liang, H.H, Lin, Z.H, Fan, Y.L, Ye, K, Gao, H.G, Tao, Y.J, Jin, M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A bacterial transcription activator dedicated to the expression of the enzyme catalyzing the first committed step in fatty acid biosynthesis.
Nucleic Acids Res., 52, 2024
|
|
5YJH
 
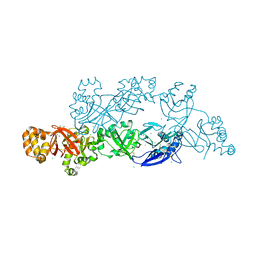 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
5YJG
 
 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYSTEINE, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
4KNG
 
 | | Crystal structure of human LGR5-RSPO1-RNF43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, E3 ubiquitin-protein ligase RNF43, Leucine-rich repeat-containing G-protein coupled receptor 5, ... | | Authors: | Chen, P.H, He, X. | | Deposit date: | 2013-05-09 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis of R-spondin recognition by LGR5 and RNF43.
Genes Dev., 27, 2013
|
|
9BVP
 
 | | Vitamin K-dependent gamma-carboxylase with TMG2 propeptide and glutamate-rich region and with vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
9BVO
 
 | | Vitamin K-dependent gamma-carboxylase in apo state | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
9BVL
 
 | | Vitamin K-dependent gamma-carboxylase with factor X propeptide and glutamate-rich region and with vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
9BVR
 
 | | Vitamin K-dependent gamma-carboxylase with factor IX propeptide and partially carboxylated glutamate-rich region and with vitamin K hydroquinone and calcium | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
9BVQ
 
 | | Vitamin K-dependent gamma-carboxylase with TMG2 propeptide and glutamate-rich region | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
9BVK
 
 | | Vitamin K-dependent gamma-carboxylase with factor IX propeptide and glutamate-rich region and with vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
9BVM
 
 | | Vitamin K-dependent gamma-carboxylase with protein C propeptide and glutamate-rich region and with vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
4KVN
 
 | | Crystal structure of Fab 39.29 in complex with Influenza Hemagglutinin A/Perth/16/2009 (H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Fong, R, Swem, L.R, Lupardus, P.J. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Novel In vivo Human Plasmablast Enrichment Technique Allows Rapid Identification of Therapeutic Anti-Influenza A Antibodies
Cell Host Microbe, 14, 2013
|
|
5K7K
 
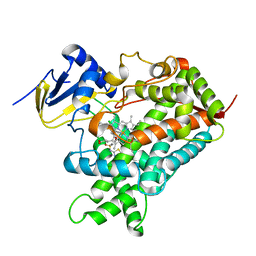 | |
7Y8Y
 
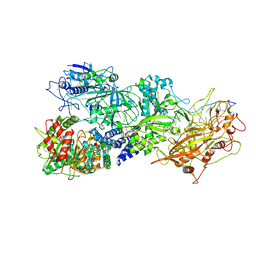 | | Structure of Cas7-11-crRNA-tgRNA in complex with TPR-CHAT | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (37-MER), ... | | Authors: | Wang, S, Guo, M, Zhu, Y, Huang, Z. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the type III-E CRISPR-Cas effector gRAMP in complex with TPR-CHAT.
Cell Res., 32, 2022
|
|
7XSC
 
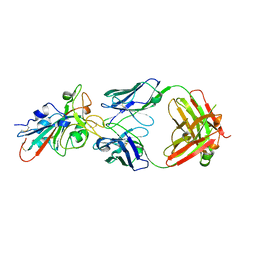 | |
5JJU
 
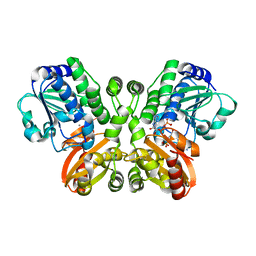 | | Crystal structure of Rv2837c complexed with 5'-pApA and 5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA (5'-R(P*AP*A)-3'), ... | | Authors: | Wang, F, He, Q, Liu, S, Gu, L. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
1B4W
 
 | |
6LPV
 
 | |
1A2A
 
 | | AGKISTROTOXIN, A PHOSPHOLIPASE A2-TYPE PRESYNAPTIC NEUROTOXIN FROM AGKISTRODON HALYS PALLAS | | Descriptor: | CHLORIDE ION, PHOSPHOLIPASE A2 | | Authors: | Tang, L, Zhou, Y, Lin, Z. | | Deposit date: | 1997-12-25 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of agkistrodotoxin, a phospholipase A2-type presynaptic neurotoxin from agkistrodon halys pallas.
J.Mol.Biol., 282, 1998
|
|
4ZRI
 
 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
