3NMZ
 
 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
3JAB
 
 | | Domain organization and conformational plasticity of the G protein effector, PDE6 | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, GafA domain of cone phosphodiesterase 6C, GafB domain of phosphodiesterase 2A, ... | | Authors: | Zhang, Z, He, F, Constantine, R, Baker, M.L, Baehr, W, Schmid, M.F, Wensel, T.G, Agosto, M.A. | | Deposit date: | 2015-05-26 | | Release date: | 2015-06-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6.
J.Biol.Chem., 290, 2015
|
|
3NMX
 
 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
1ZUC
 
 | | Progesterone receptor ligand binding domain in complex with the nonsteroidal agonist tanaproget | | Descriptor: | 5-(4,4-DIMETHYL-2-THIOXO-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-6-YL)-1-METHYL-1H-PYRROLE-2-CARBONITRILE, Progesterone receptor, SULFATE ION | | Authors: | Zhang, Z, Olland, A.M, Zhu, Y, Cohen, J, Berrodin, T, Chippari, S, Appavu, C, Li, S, Wilhem, J, Chopra, R, Fensome, A, Zhang, P, Wrobel, J, Unwalla, R.J, Lyttle, C.R, Winneker, R.C. | | Deposit date: | 2005-05-30 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and pharmacological properties of a potent and selective novel nonsteroidal progesterone receptor agonist tanaproget
J.Biol.Chem., 280, 2005
|
|
3TBL
 
 | | Structure of Mono-ubiquitinated PCNA: Implications for DNA Polymerase Switching and Okazaki Fragment Maturation | | Descriptor: | Proliferating cell nuclear antigen, Ubiquitin | | Authors: | Zhang, Z, Lee, M, Lee, E, Zhang, S. | | Deposit date: | 2011-08-07 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure of monoubiquitinated PCNA: Implications for DNA polymerase switching and Okazaki fragment maturation.
Cell Cycle, 11, 2012
|
|
7DVA
 
 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DUP
 
 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
3KCM
 
 | |
3KXW
 
 | |
3NPK
 
 | |
3KZB
 
 | |
3L0Q
 
 | |
3OU8
 
 | |
3LIF
 
 | |
3OTT
 
 | |
3LIE
 
 | |
3PAN
 
 | |
3LIC
 
 | |
3LIB
 
 | |
3LID
 
 | |
3OPN
 
 | |
3LI9
 
 | |
3LI8
 
 | |
3LNV
 
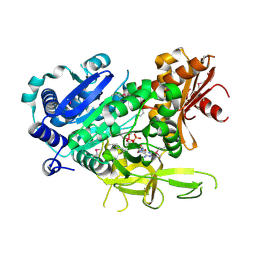 | | The crystal structure of fatty acyl-adenylate ligase from L. pneumophila in complex with acyl adenylate and pyrophosphate | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, PYROPHOSPHATE 2-, Saframycin Mx1 synthetase B | | Authors: | Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-03 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies of Fatty Acyl Adenylate Ligases from E. coli and L. pneumophila.
J.Mol.Biol., 406, 2011
|
|
