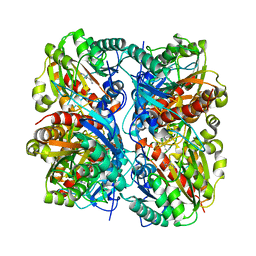
1VDD
 
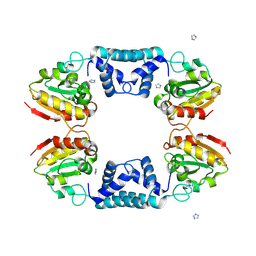 | |
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
1UMF
 
 | |
1TAQ
 
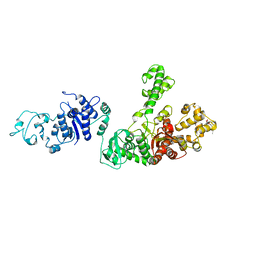 | | STRUCTURE OF TAQ DNA POLYMERASE | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, TAQ DNA POLYMERASE, ZINC ION | | Authors: | Kim, Y, Eom, S.H, Wang, J, Lee, D.-S, Suh, S.W, Steitz, T.A. | | Deposit date: | 1996-06-04 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus aquaticus DNA polymerase.
Nature, 376, 1995
|
|
1WOR
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, DIHYDROLIPOIC ACID | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
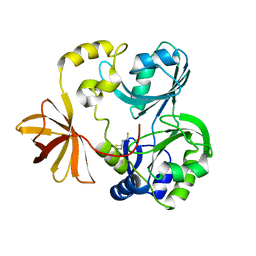
1WOG
 
 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | HEXANE-1,6-DIAMINE, MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1UM0
 
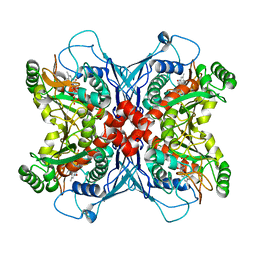 | | Crystal structure of chorismate synthase complexed with FMN | | Descriptor: | Chorismate synthase, FLAVIN MONONUCLEOTIDE | | Authors: | Ahn, H.J, Yoon, H.J, Lee, B, Suh, S.W. | | Deposit date: | 2003-09-18 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of chorismate synthase: a novel FMN-binding protein fold and functional insights
J.Mol.Biol., 336, 2004
|
|
1VC1
 
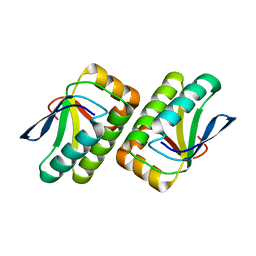 | | Crystal structure of the TM1442 protein from Thermotoga maritima, a homolog of the Bacillus subtilis general stress response anti-anti-sigma factor RsbV | | Descriptor: | Putative anti-sigma factor antagonist TM1442 | | Authors: | Lee, J.Y, Ahn, H.J, Ha, K.S, Suh, S.W. | | Deposit date: | 2004-03-03 | | Release date: | 2004-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the TM1442 protein from Thermotoga maritima, a homolog of the Bacillus subtilis general stress response anti-anti-sigma factor RsbV
Proteins, 56, 2004
|
|
1WOH
 
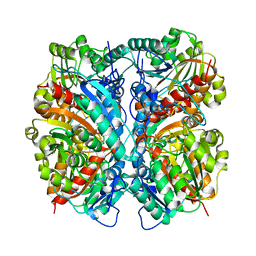 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1WOI
 
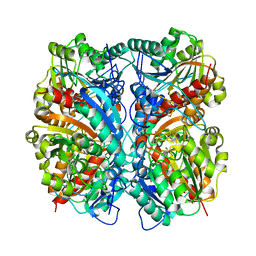 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1WOP
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
1WOS
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
1WOO
 
 | | Crystal structure of T-protein of the Glycine Cleavage System | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Aminomethyltransferase | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
1WUW
 
 | | Crystal Structure of beta hordothionin | | Descriptor: | Beta-hordothionin, PARA-TOLUENE SULFONATE, SERINE | | Authors: | Johnson, K.A, Kim, E, Teeter, M.M, Suh, S.W, Stec, B. | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alpha-hordothionin at 1.9 Angstrom resolution.
Febs Lett., 579, 2005
|
|
1ECY
 
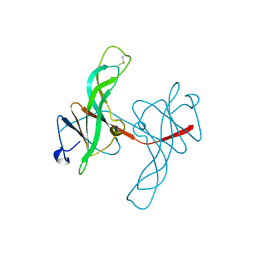 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1EK8
 
 | |
1ECZ
 
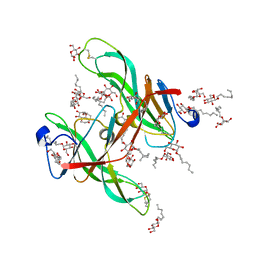 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, octyl beta-D-glucopyranoside | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1NP3
 
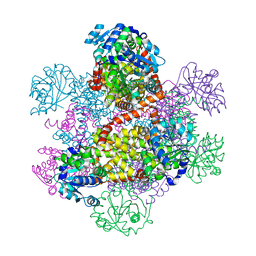 | | Crystal structure of class I acetohydroxy acid isomeroreductase from Pseudomonas aeruginosa | | Descriptor: | Ketol-acid reductoisomerase | | Authors: | Ahn, H.J, Eom, S.J, Yoon, H.-J, Lee, B.I, Cho, H, Suh, S.W. | | Deposit date: | 2003-01-17 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Class I Acetohydroxy Acid Isomeroreductase from Pseudomonas aeruginosa
J.Mol.Biol., 328, 2003
|
|
1PX8
 
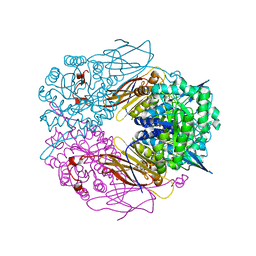 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1Q1Y
 
 | | Crystal Structures of Peptide Deformylase from Staphylococcus aureus Complexed with Actinonin | | Descriptor: | ACTINONIN, Peptide deformylase, ZINC ION | | Authors: | Yoon, H.J, Lee, S.K, Kim, H.L, Kim, H.W, Kim, H.W, Lee, J.Y, Mikami, B, Suh, S.W. | | Deposit date: | 2003-07-23 | | Release date: | 2004-07-23 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
1YUN
 
 | | Crystal Structure of Nicotinic Acid Mononucleotide Adenylyltransferase from Pseudomonas aeruginosa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable nicotinate-nucleotide adenylyltransferase | | Authors: | Yoon, H.J, Kim, H.L, Mikami, B, Suh, S.W. | | Deposit date: | 2005-02-14 | | Release date: | 2005-11-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of nicotinic acid mononucleotide adenylyltransferase from Pseudomonas aeruginosa in its Apo and substrate-complexed forms reveals a fully open conformation
J.Mol.Biol., 351, 2005
|
|
1YUM
 
 | | Crystal Structure of Nicotinic Acid Mononucleotide Adenylyltransferase from Pseudomonas aeruginosa | | Descriptor: | 'Probable nicotinate-nucleotide adenylyltransferase, CITRIC ACID, NICOTINATE MONONUCLEOTIDE | | Authors: | Yoon, H.J, Kim, H.L, Mikami, B, Suh, S.W. | | Deposit date: | 2005-02-14 | | Release date: | 2005-11-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of nicotinic acid mononucleotide adenylyltransferase from Pseudomonas aeruginosa in its Apo and substrate-complexed forms reveals a fully open conformation
J.Mol.Biol., 351, 2005
|
|
1YGZ
 
 | | Crystal Structure of Inorganic Pyrophosphatase from Helicobacter pylori | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Wu, C.A, Lokanath, N.K, Kim, D.Y, Park, H.J, Hwang, H.Y, Kim, S.T, Suh, S.W, Kim, K.K. | | Deposit date: | 2005-01-06 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of inorganic pyrophosphatase from Helicobacter pylori.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2ARU
 
 | | Crystal structure of lipoate-protein ligase A bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
2ARS
 
 | | Crystal structure of lipoate-protein ligase A From Thermoplasma acidophilum | | Descriptor: | Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
