6MZI
 
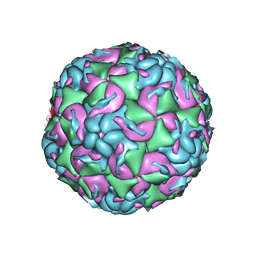 | | CryoEM structure of human enterovirus D68 expanded 1 particle (pH 6.5, 4 degrees Celsius, 3 min) | | Descriptor: | viral protein 1, viral protein 2, viral protein 3, ... | | Authors: | Liu, Y, Rossmann, M.G. | | Deposit date: | 2018-11-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Molecular basis for the acid-initiated uncoating of human enterovirus D68.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6D8A
 
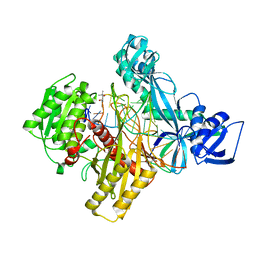 | | RsAgo Ternary Complex with guide RNA and Target DNA Containing A-A Bulge Within the Seed Segment of the Target Strand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*AP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
8T4Q
 
 | |
3BPW
 
 | |
7N8N
 
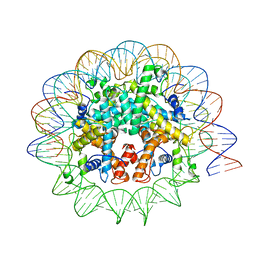 | | Melbournevirus nucleosome like particle | | Descriptor: | DNA (147-MER), Histone H2B-H2A doublet, Histone H4-H3 doublet | | Authors: | Liu, Y, Toner, C.M, Zhou, K, Bowerman, S, Luger, K. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Virus-encoded histone doublets are essential and form nucleosome-like structures.
Cell, 184, 2021
|
|
5BNO
 
 | | Crystal structure of human enterovirus D68 in complex with 6'SLN | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Sheng, J, Meng, G, Xiao, C, Rossmann, M.G. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Sialic acid-dependent cell entry of human enterovirus D68.
Nat Commun, 6, 2015
|
|
3BGJ
 
 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 6-iodo-UMP | | Descriptor: | GLYCEROL, URIDINE-5'-MONOPHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Bello, A.M, Devalla, S, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-26 | | Release date: | 2008-11-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 6-iodo-UMP
To be Published
|
|
3BGG
 
 | | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Wang, X.Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP
To be Published
|
|
5Z5K
 
 | | Structure of the DCC-Draxin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Draxin, ... | | Authors: | Liu, Y, Xiao, J, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC.
Neuron, 97, 2018
|
|
5BNN
 
 | | Crystal structure of human enterovirus D68 in complex with 6'SL | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Sheng, J, Meng, G, Xiao, C, Rossmann, M.G. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Sialic acid-dependent cell entry of human enterovirus D68.
Nat Commun, 6, 2015
|
|
5CQ2
 
 | | Crystal Structure of tandem WW domains of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the regulatory role of the PPxY motifs in the thioredoxin-interacting protein TXNIP.
Biochem.J., 473, 2016
|
|
3BVJ
 
 | |
2ESH
 
 | | Crystal Structure of Conserved Protein of Unknown Function TM0937- a Potential Transcriptional Factor | | Descriptor: | CALCIUM ION, conserved hypothetical protein TM0937 | | Authors: | Liu, Y, Bochkareva, E, Zheng, H, Xu, X, Nocek, B, Lunin, V, Edward, A, Pai, E.F, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein TM0937
To be Published
|
|
2FRH
 
 | | Crystal Structure of Sara, A Transcription Regulator From Staphylococcus Aureus | | Descriptor: | CALCIUM ION, Staphylococcal accessory regulator A | | Authors: | Liu, Y, Manna, A.C, Ingavale, S, Cheung, A.L, Zhang, G. | | Deposit date: | 2006-01-19 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
9DLF
 
 | | Arabinosyltransferase AftB in complex with Fab_B3 | | Descriptor: | Arabinosyltransferase AftB, Fab_B3 heavy chain, Fab_B3 light chain, ... | | Authors: | Liu, Y, Mancia, F. | | Deposit date: | 2024-09-10 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
9DM5
 
 | | Product-Bound mannosyltransferase PimE | | Descriptor: | (2S)-1-{[(S)-{[(1S,2R,3R,4S,5S,6R)-2-[(6-O-hexadecanoyl-beta-L-gulopyranosyl)oxy]-3,4,5-trihydroxy-6-{[beta-D-mannopyranosyl-(1->2)-alpha-D-mannopyranosyl-(1->6)-beta-D-mannopyranosyl-(1->6)-alpha-D-mannopyranosyl]oxy}cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}-3-(hexadecanoyloxy)propan-2-yl 10-methyloctadecanoate, MONO-TRANS, OCTA-CIS DECAPRENYL-PHOSPHATE, ... | | Authors: | Liu, Y. | | Deposit date: | 2024-09-12 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
9DM7
 
 | | mannosyltransferase PimE in complex with Fab_E6 | | Descriptor: | Fab_E6 heavy chain, Fab_E6 light chain, Mannosyltransferase | | Authors: | Liu, Y. | | Deposit date: | 2024-09-12 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
2FNP
 
 | | Crystal structure of SarA | | Descriptor: | Staphylococcal accessory regulator A | | Authors: | Liu, Y, Manna, A.C, Pan, C.H, Cheung, A.L, Zhang, G. | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
9F5H
 
 | | Crystal structure of MGAT5 bump-and-hole mutant in complex with UDP and M592 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Secreted alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Liu, Y, Bineva-Todd, G, Meek, R, Mazo, L, Piniello, B, Moroz, O.V, Begum, N, Roustan, C, Tomita, S, Kjaer, S, Rovira, C, Davies, G.J, Schumann, B. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Bioorthogonal Precision Tool for Human N -Acetylglucosaminyltransferase V.
J.Am.Chem.Soc., 146, 2024
|
|
2GJ2
 
 | | Crystal Structure of VP9 from White Spot Syndrome Virus | | Descriptor: | CADMIUM ION, wsv230 | | Authors: | Liu, Y, Wu, J.L, Song, J.X, Sivaraman, J, Hew, C.L. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a Novel Nonstructural Protein, VP9, from White Spot Syndrome Virus: Its Structure Reveals a Ferredoxin Fold with Specific Metal Binding Sites
J.Virol., 80, 2006
|
|
2GJI
 
 | | NMR solution structure of VP9 from White Spot Syndrome Virus | | Descriptor: | wsv230 | | Authors: | Liu, Y, Wu, J.L, Song, J.X, Sivaraman, J, Hew, C.L. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Identification of a Novel Nonstructural Protein, VP9, from White Spot Syndrome Virus: Its Structure Reveals a Ferredoxin Fold with Specific Metal Binding Sites
J.Virol., 80, 2006
|
|
2EAW
 
 | |
8JFQ
 
 | | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-back Loop | | Descriptor: | 26mer-DNA | | Authors: | Liu, Y, Li, J, Zhang, Y, Wang, Y, Chen, J, Bian, Y, Xia, Y, Yang, M.H, Zheng, K, Wang, K.B, Kong, L.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-Back Loop.
J.Am.Chem.Soc., 145, 2023
|
|
3Q9S
 
 | |
3PSP
 
 | |
