3PZJ
 
 | | Crystal structure of a probable acetyltransferases (GNAT family) from Chromobacterium violaceum ATCC 12472 | | Descriptor: | 1,2-ETHANEDIOL, Probable acetyltransferases, SODIUM ION | | Authors: | Nocek, B, Stein, A, Bigelow, L, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-14 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a probable acetyltransferases (GNAT family) from Chromobacterium violaceum ATCC 12472
TO BE PUBLISHED
|
|
3SOY
 
 | | Nuclear transport factor 2 (NTF2-like) superfamily protein from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | GLYCEROL, IMIDAZOLE, MALONATE ION, ... | | Authors: | Cuff, M.E, Li, H, Jedrzejczak, R, Brown, R.N, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nuclear transport factor 2 (NTF2-like) superfamily protein from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
TO BE PUBLISHED
|
|
3T6O
 
 | | The Structure of an Anti-sigma-factor antagonist (STAS) domain protein from Planctomyces limnophilus. | | Descriptor: | CHLORIDE ION, Sulfate transporter/antisigma-factor antagonist STAS | | Authors: | Cuff, M.E, Moser, C, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of an Anti-sigma-factor antagonist (STAS) domain protein from Planctomyces limnophilus.
TO BE PUBLISHED
|
|
1NG5
 
 | |
3V75
 
 | | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680 | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Stogios, P.J, Xu, X, Cui, H, Kudritska, M, Tan, K, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680
To be Published
|
|
2QMM
 
 | |
3UO3
 
 | | Jac1 co-chaperone from Saccharomyces cerevisiae, 5-182 clone | | Descriptor: | ACETATE ION, J-type co-chaperone JAC1, mitochondrial | | Authors: | Osipiuk, J, Bigelow, L, Mulligan, R, Feldmann, B, Babnigg, G, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
3O6P
 
 | | Crystal structure of peptide ABC transporter, peptide-binding protein | | Descriptor: | Peptide ABC transporter, peptide-binding protein, SODIUM ION | | Authors: | Chang, C, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of peptide ABC transporter, peptide-binding protein
To be Published
|
|
3NZR
 
 | |
3O66
 
 | | Crystal structure of glycine betaine/carnitine/choline ABC transporter | | Descriptor: | ACETATE ION, Glycine betaine/carnitine/choline ABC transporter, TRIETHYLENE GLYCOL | | Authors: | Chang, C, Bigelow, L, Carroll, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of glycine betaine/carnitine/choline ABC transporter
To be Published
|
|
3OM8
 
 | | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable hydrolase | | Authors: | Tan, K, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01
To be Published
|
|
3QWU
 
 | | Putative ATP-dependent DNA ligase from Aquifex aeolicus. | | Descriptor: | ADENOSINE, CALCIUM ION, DNA ligase, ... | | Authors: | Osipiuk, J, Quartey, P, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-28 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Putative ATP-dependent DNA ligase from Aquifex aeolicus.
To be Published
|
|
3QSJ
 
 | | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius | | Descriptor: | CALCIUM ION, GLYCEROL, NUDIX hydrolase | | Authors: | Michalska, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius
To be Published
|
|
3QQZ
 
 | | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein yjiK | | Authors: | Stein, A, Chhor, G, Nocek, B, Fenske, R.J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073
TO BE PUBLISHED
|
|
1I6N
 
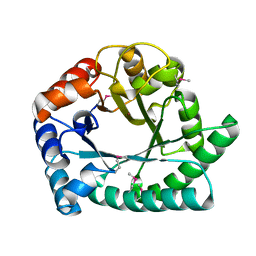 | | 1.8 A Crystal structure of IOLI protein with a binding zinc atom | | Descriptor: | IOLI PROTEIN, ZINC ION | | Authors: | Zhang, R.G, Dementiva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Alkire, R, Maltsev, N, Korolev, O, Dieckman, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-03-02 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
3NZE
 
 | | The crystal structure of a domain of a possible sugar-binding transcriptional regulator from Arthrobacter aurescens TC1. | | Descriptor: | CALCIUM ION, Putative transcriptional regulator, sugar-binding family | | Authors: | Tan, K, Zhang, R, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The crystal structure of a domain of a possible sugar-binding transcriptional regulator from Arthrobacter aurescens TC1.
To be Published
|
|
3QZ6
 
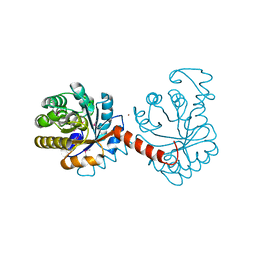 | |
5SUJ
 
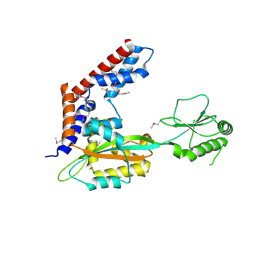 | | Crystal structure of uncharacterized protein LPG2148 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Discovery of Ubiquitin Deamidases in the Pathogenic Arsenal of Legionella pneumophila.
Cell Rep, 23, 2018
|
|
3OOV
 
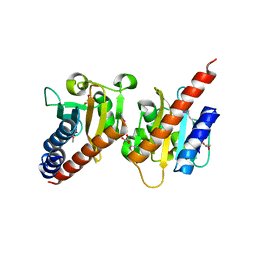 | | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287 | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, putative | | Authors: | Joachimiak, A, Duke, N.E.C, Hatzos-Skintges, C, Mulligan, R, Clancy, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287
To be Published
|
|
1NSL
 
 | | Crystal structure of Probable acetyltransferase | | Descriptor: | CHLORIDE ION, Probable acetyltransferase | | Authors: | Brunzelle, J.S, Korolev, S.V, Wu, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YdaF protein: A putative ribosomal N-acetyltransferase
Proteins, 57, 2004
|
|
3P9Z
 
 | | Crystal structure of uroporphyrinogen-III synthetase from Helicobacter pylori 26695 | | Descriptor: | MALONATE ION, Uroporphyrinogen III cosynthase (HemD) | | Authors: | Nocek, B, Stein, A, Chhor, G, Fenske, R.J, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-18 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uroporphyrinogen-III synthetase from Helicobacter pylori 26695
To be Published
|
|
3RRL
 
 | | Complex structure of 3-oxoadipate coA-transferase subunit A and B from Helicobacter pylori 26695 | | Descriptor: | GLYCEROL, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit A, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit B | | Authors: | Nocek, B, Stein, A, Marshall, N, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Complex structure of 3-oxoadipate coA-transferase subunit A and B
from Helicobacter pylori 26695
TO BE PUBLISHED
|
|
5TSC
 
 | | The crystal structure of Lpg2147 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Valleau, D, Xu, X, Cui, H, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | The crystal structure of Lpg2147 from Legionella pneumophila
To Be Published
|
|
3QOC
 
 | | Crystal structure of N-terminal domain (Creatinase/Prolidase like domain) of putative metallopeptidase from Corynebacterium diphtheriae | | Descriptor: | CHLORIDE ION, Putative metallopeptidase, SULFATE ION | | Authors: | Nocek, B, Stein, A, Marshall, N, Putagunta, R, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-09 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of N-terminal domain (Creatinase/Prolidase like domain) of putative metallopeptidase from Corynebacterium diphtheriae
TO BE PUBLISHED
|
|
3UO2
 
 | | Jac1 co-chaperone from Saccharomyces cerevisiae | | Descriptor: | J-type co-chaperone JAC1, mitochondrial | | Authors: | Osipiuk, J, Mulligan, R, Bigelow, L, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
