3E0X
 
 | |
3D1P
 
 | | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative thiosulfate sulfurtransferase YOR285W | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae.
To be Published
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
3CNG
 
 | | Crystal structure of NUDIX hydrolase from Nitrosomonas europaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
3NA2
 
 | | Crystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crsystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum
To be Published
|
|
2RFQ
 
 | | Crystal structure of 3-HSA hydroxylase from Rhodococcus sp. RHA1 | | Descriptor: | 3-HSA hydroxylase, oxygenase, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE | | Authors: | Chang, C, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-01 | | Release date: | 2007-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of 3-HSA hydroxylase, oxygenase from Rhodococcus sp. RHA1.
To be Published
|
|
2RA5
 
 | | Crystal structure of the putative transcriptional regulator from Streptomyces coelicolor | | Descriptor: | ISOPROPYL ALCOHOL, Putative transcriptional regulator, S,R MESO-TARTARIC ACID | | Authors: | Kim, Y, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the putative transcriptional regulator from Streptomyces coelicolor.
To be Published
|
|
2RAE
 
 | | Crystal structure of a TetR/AcrR family transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | Transcriptional regulator, AcrR family protein | | Authors: | Zhang, R, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a TetR/AcrR family transcriptional regulator from Rhodococcus sp. RHA1
To be Published
|
|
2RBC
 
 | | Crystal structure of a putative ribokinase from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-11-13 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a putative ribokinase from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
2R6I
 
 | | Crystal structure of Atu1473 protein, a putative chaperone from Agrobacterium tumefaciens | | Descriptor: | Uncharacterized protein Atu1473 | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of Atu1473 protein, a putative chaperone from Agrobacterium tumefaciens.
To be Published
|
|
2R6U
 
 | | Crystal structure of gene product RHA04853 from Rhodococcus sp. RHA1 | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-06 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of gene product RHA04853 from Rhodococcus sp. RHA1.
To be Published
|
|
3E6Q
 
 | | Putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure of putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa.
To be Published
|
|
2P7J
 
 | |
3E8X
 
 | | Putative NAD-dependent epimerase/dehydratase from Bacillus halodurans. | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative NAD-dependent epimerase/dehydratase | | Authors: | Osipiuk, J, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of putative NAD-dependent epimerase/dehydratase from Bacillus halodurans.
To be Published
|
|
2HWJ
 
 | | Crystal structure of protein Atu1540 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu1540 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-01 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of protein Atu1540 from Agrobacterium tumefaciens
To be Published
|
|
2HXO
 
 | | Structure of the transcriptional regulator SCO7222, a TetR from Streptomyces coelicolor | | Descriptor: | Putative TetR-family transcriptional regulator | | Authors: | Singer, A.U, Skarina, T, Zhang, R.G, Onopriyenko, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transcriptional regulator SCO7222, a TetR from Streptomyces coelicolor
To be Published
|
|
4PF1
 
 | | Crystal structure of aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon | | Descriptor: | GLYCEROL, Peptidase S15/CocE/NonD, TRIETHYLENE GLYCOL | | Authors: | Michalska, K, Chhor, G, Fayman, K, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New aminopeptidase from "microbial dark matter" archaeon.
FASEB J., 29, 2015
|
|
1ZFJ
 
 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
2FU2
 
 | | Crystal structure of protein SPy2152 from Streptococcus pyogenes | | Descriptor: | Hypothetical protein SPy2152 | | Authors: | Chang, C, Cymborowski, M, Otwinowski, Z, Minor, W, Lezondra, L.-E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-25 | | Release date: | 2006-03-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of pyogenecin immunity protein, a novel bacteriocin-like immunity protein from Streptococcus pyogenes.
Bmc Struct.Biol., 9, 2009
|
|
2FCK
 
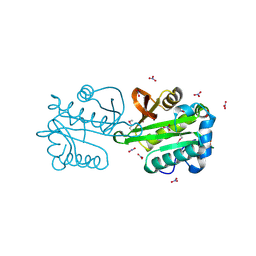 | | Structure of a putative ribosomal-protein-serine acetyltransferase from Vibrio cholerae. | | Descriptor: | GLYCEROL, NITRATE ION, ribosomal-protein-serine acetyltransferase, ... | | Authors: | Cuff, M.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-28 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an acetyltransferase protein from Vibrio cholerae strain N16961.
Proteins, 69, 2007
|
|
3QOD
 
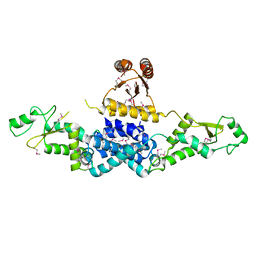 | | Crystal Structure of Heterocyst Differentiation Protein, HetR from Fischerella mv11 | | Descriptor: | Heterocyst differentiation protein | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure of transcription factor HetR required for heterocyst differentiation in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QOE
 
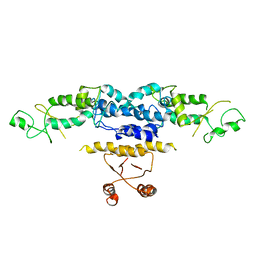 | | Crystal Structure of Heterocyst Differentiation Protein, HetR from Fischerella mv11 | | Descriptor: | Heterocyst differentiation protein | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structure of transcription factor HetR required for heterocyst differentiation in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3F3K
 
 | | The structure of uncharacterized protein YKR043C from Saccharomyces cerevisiae. | | Descriptor: | GLYCEROL, Uncharacterized protein YKR043C | | Authors: | Cuff, M, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
3HL0
 
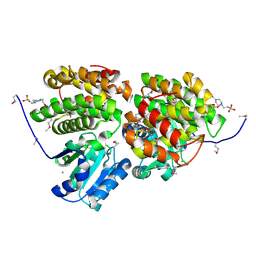 | | Crystal structure of Maleylacetate reductase from Agrobacterium tumefaciens | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Maleylacetate reductase, ... | | Authors: | Chang, C, Evdokimova, E, Mursleen, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Maleylacetate reductase from Agrobacterium tumefaciens
To be Published
|
|
3GHD
 
 | | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Descriptor: | a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Authors: | Dong, A, Xu, X, Chruszcz, M, Brown, G, Proudfoot, M, Edwards, A.M, Joachimiak, A, Minor, W, Savchenko, A, Yaleunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain
To be Published
|
|
