4GPW
 
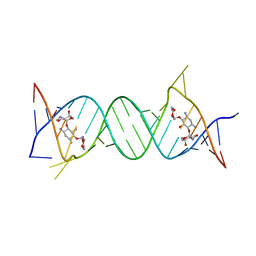 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-hydroxysisomicin (P21212 form) | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
1YBD
 
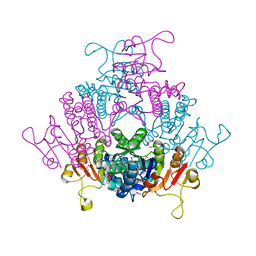 | |
4GPY
 
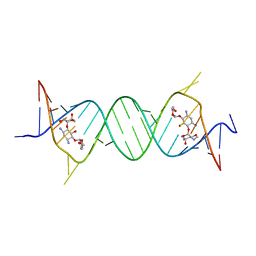 | | Crystal structure of the bacterial ribosomal decoding site in complex with 6'-hydroxysisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
1Y9I
 
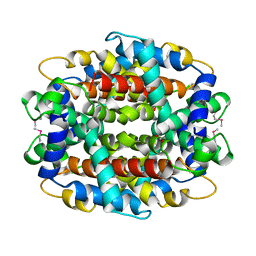 | | Crystal structure of low temperature requirement C protein from Listeria monocytogenes | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphatidylglycerophosphatase (PGPase), a putative membrane-bound lipid phosphatase, reveals a novel binuclear metal binding site and two "proton wires".
Proteins, 64, 2006
|
|
3V4C
 
 | | Crystal structure of a semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Aldehyde dehydrogenase (NADP+) | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of a semialdehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3US8
 
 | | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Isocitrate dehydrogenase [NADP], SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
7YK5
 
 | | Rubisco from Phaeodactylum tricornutum bound to PYCO1(452-592) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, Multifunctional fusion protein, PYCO1 LSU binding motif, ... | | Authors: | Oh, Z.G, Ang, W.S.L, Bhushan, S, Mueller-Cajar, O. | | Deposit date: | 2022-07-21 | | Release date: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | A linker protein from a red-type pyrenoid phase separates with Rubisco via oligomerizing sticker motifs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2POF
 
 | |
2PUZ
 
 | | Crystal structure of Imidazolonepropionase from Agrobacterium tumefaciens with bound product N-formimino-L-Glutamate | | Descriptor: | CHLORIDE ION, FE (III) ION, Imidazolonepropionase, ... | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution.
Proteins, 69, 2007
|
|
1XD7
 
 | |
2NRJ
 
 | | Crystal Structure of Hemolysin binding component from Bacillus cereus | | Descriptor: | Hbl B protein | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the B component of Hemolysin BL from Bacillus cereus
Proteins, 71, 2008
|
|
7XW7
 
 | | TSHR-K1-70 complex | | Descriptor: | K1-70 scFv, Thyrotropin receptor | | Authors: | Duan, J, Xu, P, Luan, X, Ji, Y, Yuan, Q, He, X, Ye, J, Cheng, X, Jiang, H, Zhang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-05-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Hormone- and antibody-mediated activation of the thyrotropin receptor.
Nature, 609, 2022
|
|
3URH
 
 | | Crystal structure of a dihydrolipoamide dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | 1,2-ETHANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a dihydrolipoamide dehydrogenase from Sinorhizobium meliloti 1021
TO BE PUBLISHED
|
|
2PHP
 
 | |
2XXA
 
 | | The Crystal Structure of the Signal Recognition Particle (SRP) in Complex with its Receptor(SR) | | Descriptor: | 4.5S RNA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Ataide, S.F, Schmitz, N, Shen, K, Ke, A, Shan, S, Doudna, J.A, Ban, N. | | Deposit date: | 2010-11-09 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | The Crystal Structure of the Signal Recognition Particle in Complex with its Receptor.
Science, 331, 2011
|
|
4I0X
 
 | | Crystal structure of the Mycobacterum abscessus EsxEF (Mab_3112-Mab_3113) complex | | Descriptor: | BETA-MERCAPTOETHANOL, ESAT-6-like protein MAB_3112, ESAT-6-like protein MAB_3113, ... | | Authors: | Arbing, M.A, Chan, S, Lu, J, Kuo, E, Harris, L, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-11-19 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
2Q09
 
 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
1YIR
 
 | |
1YAV
 
 | |
1YBE
 
 | |
1BF4
 
 | | CHROMOSOMAL DNA-BINDING PROTEIN SSO7D/D(GCGAACGC) COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*5IUP*CP*GP*C)-3'), PROTEIN (CHROMOSOMAL PROTEIN SSO7D) | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Padmanabhan, S, Lim, L, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1998-05-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.
Nat.Struct.Biol., 5, 1998
|
|
7XW6
 
 | | TSHR-Gs-M22 antibody-ML109 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Duan, J, Xu, P, Luan, X, Ji, Y, Yuan, Q, He, X, Ye, J, Cheng, X, Jiang, H, Zhang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-05-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Hormone- and antibody-mediated activation of the thyrotropin receptor.
Nature, 609, 2022
|
|
2QQ6
 
 | |
3WR1
 
 | | Crystal structure of Cormorant (Phalacrocorax carbo) hemoglobin | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jagadeesan, G, Vinodh Kumar, V, Peters, H.G, Malathy, P, Harikrishna Etti, S, Gunasekaran, K, Aravindhan, S. | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of Cormorant (Phalacrocorax carbo) hemoglobin
To be Published
|
|
2X6M
 
 | | Structure of a single domain camelid antibody fragment in complex with a C-terminal peptide of alpha-synuclein | | Descriptor: | ALPHA-SYNUCLEIN PEPTIDE, HEAVY CHAIN VARIABLE DOMAIN FROM DROMEDARY | | Authors: | DeGenst, E, Guilliams, T, Wellens, J, O'Day, E.M, Waudby, C.A, Meehan, S, Dumoulin, M, Hsu, S.-T.D, Cremades, N, Verschueren, K.H.G, Pardon, E, Wyns, L, Steyaert, J, Christodoulou, J, Dobson, C.M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Properties of a Complex of Alpha-Synuclein and a Single-Domain Camelid Antibody.
J.Mol.Biol., 402, 2010
|
|
