5K08
 
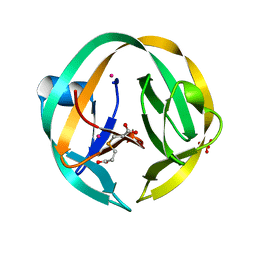 | | RecA mini intein-Zeise's salt complex | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, RecA mini intein, SULFATE ION, ... | | Authors: | Li, Z, Li, H.M. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural insights into protein splicing inhibition by platinum therapeutics as potential anti-microbials
To be published
|
|
4AGW
 
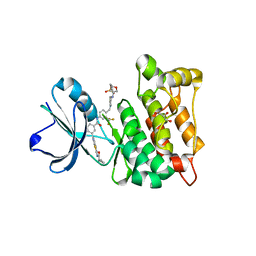 | | Discovery of a small molecule type II inhibitor of wild-type and gatekeeper mutants of BCR-ABL, PDGFRalpha, Kit, and Src kinases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-{2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromet hyl)phenyl}benzamide, GLYCEROL, ... | | Authors: | Weisberg, E, Choi, H.G, Seeliger, M, Gray, N, Griffin, J.D. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Small-Molecule Type II Inhibitor of Wild-Type and Gatekeeper Mutants of Bcr-Abl, Pdgfralpha, Kit, and Src Kinases: Novel Type II Inhibitor of Gatekeeper Mutants.
Blood, 115, 2010
|
|
7DTK
 
 | |
7DTJ
 
 | |
3C0C
 
 | |
6WQL
 
 | |
6WQJ
 
 | |
3RL7
 
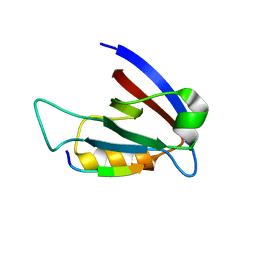 | | Crystal structure of hDLG1-PDZ1 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
3RES
 
 | |
3KU0
 
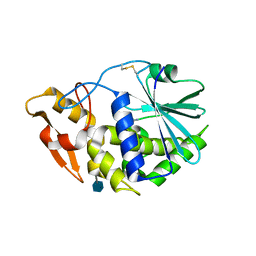 | | Structure of GAP31 with adenine at its binding pocket | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3SRQ
 
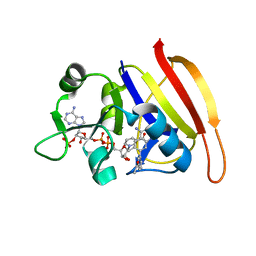 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 1-[3-(2,4-diamino-6-methylquinazolin-7-yl)phenyl]ethanone, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4MO7
 
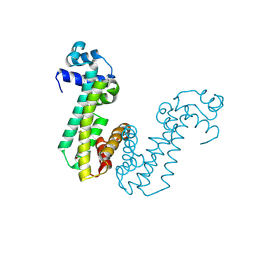 | | Crystal structure of superantigen PfiT | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Transcriptional regulator I2 | | Authors: | Liu, L.H, Chen, H, Li, H.M. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Pfit is a structurally novel Crohn's disease-associated superantigen.
Plos Pathog., 9, 2013
|
|
3SRS
 
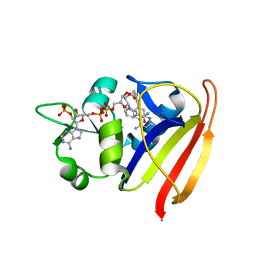 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 7-(5-bromo-2-ethoxyphenyl)-6-methylquinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3TUO
 
 | |
7XP6
 
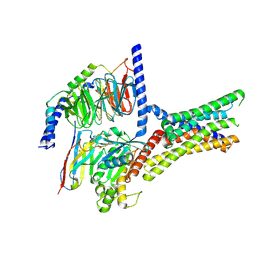 | | Cryo-EM structure of a class T GPCR in active state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP4
 
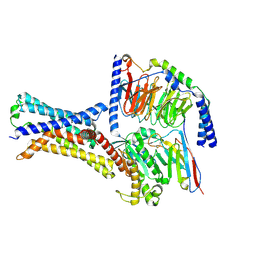 | | Cryo-EM structure of a class T GPCR in apo state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP5
 
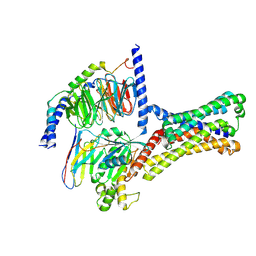 | | Cryo-EM structure of a class T GPCR in ligand-free state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
2L3T
 
 | | Solution structure of tandem SH2 domain from Spt6 | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Liu, J, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-09-22 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tandem SH2 domains from Spt6 and their binding to the phosphorylated RNA polymerase II C-terminal domain
To be Published
|
|
3KTZ
 
 | | Structure of GAP31 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2K3Q
 
 | |
2K3P
 
 | |
2NB4
 
 | | Solution structure of Q388A3 PDZ domain | | Descriptor: | Putative uncharacterized protein | | Authors: | Mei, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Q388A3 PDZ domain from Trypanosoma brucei
J.Struct.Biol., 194, 2016
|
|
2N7S
 
 | | Solution Structure of Leptospiral LigA4 Big Domain | | Descriptor: | Ig-like repeat domain protein 1 | | Authors: | Mei, S. | | Deposit date: | 2015-09-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of leptospiral LigA4 Big domain.
Biochem. Biophys. Res. Commun., 467, 2015
|
|
8I4V
 
 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
5B7W
 
 | |
