4IHA
 
 | | Crystal structure of rice DWARF14 (D14) in complex with a GR24 hydrolysis intermediate | | Descriptor: | (2R,3R)-2,4,4-trihydroxy-3-methylbutanal, Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH4
 
 | | Crystal structure of Arabidopsis DWARF14 orthologue, AtD14 | | Descriptor: | AT3g03990/T11I18_10 | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
7N0U
 
 | | Complex of recombinant Bet v 1 with Fab fragment of REGN5713 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, REGN5713 Fab fragment heavy chain, REGN5713 Fab fragment light chain, ... | | Authors: | Franklin, M.C, Romero Hernandez, A. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Targeting immunodominant Bet v 1 epitopes with monoclonal antibodies prevents the birch allergic response.
J.Allergy Clin.Immunol., 149, 2022
|
|
7N0V
 
 | |
7MXL
 
 | |
4AA4
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 22 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-METHYL-3-[6-(4-METHYLPIPERAZIN-1-YL)-4-OXIDANYLIDENE-QUINAZOLIN-3-YL]PHENYL]FURAN-3-CARBOXAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AA0
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 2 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-methyl-3-[6-(4-methylpiperazin-1-yl)-4-oxidanylidene-quinazolin-3-yl]phenyl]-2-morpholin-4-yl-pyridine-4-carboxamide | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4A9Y
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 8 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-(3-{[7-METHOXY-6-(2-PYRROLIDIN-1-YLETHOXY)QUINAZOLIN-4-YL]AMINO}-4-METHYLPHENYL)-2-MORPHOLIN-4-YLISONICOTINAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AAC
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 29 | | Descriptor: | CHLORIDE ION, MITOGEN-ACTIVATED PROTEIN KINASE 14, N-isoxazol-3-yl-4-methyl-3-[6-(4-methylpiperazin-1-yl)-4-oxo-quinazolin-3-yl]benzamide | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AA5
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 33 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-CYCLOPROPYL-4-METHYL-3-[6-(4-METHYLPIPERAZIN-1-YL)-4-OXIDANYLIDENE-QUINAZOLIN-3-YL]BENZAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-05-16 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8POA
 
 | | Structure of Coxsackievirus A16 (G-10) 2A protease | | Descriptor: | GLYCEROL, Protease 2A, ZINC ION | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-04 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of EV A71 2A protease - to be published
To Be Published
|
|
7CU2
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES ALBOGRISEOLUS FLAVIN-DEPENDENT TRYPTOPHAN 6-HALOGENASE THAL IN COMPLEX WITH REDUCED FAD | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Tryptophan 6-halogenase | | Authors: | Chitnumsub, P, Jaruwat, A, Phintha, A, Chaiyen, P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the low catalytic capability of flavin-dependent halogenases.
J.Biol.Chem., 296, 2020
|
|
7CU0
 
 | | Crystal structure of Streptomyces albogriseolus flavin-dependent tryptophan 6-halogenase Thal in complex with tryptophan | | Descriptor: | TRYPTOPHAN, Tryptophan 6-halogenase | | Authors: | Chitnumsub, P, Jaruwat, A, Phintha, A, Chaiyen, P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissecting the low catalytic capability of flavin-dependent halogenases.
J.Biol.Chem., 296, 2020
|
|
7CU1
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES ALBOGRISEOLUS FLAVIN-DEPENDENT TRYPTOPHAN 6-HALOGENASE (THAL) IN COMPLEX WITH FAD and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Tryptophan 6-halogenase | | Authors: | Chitnumsub, P, Jaruwat, A, Phintha, A, Chaiyen, P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Dissecting the low catalytic capability of flavin-dependent halogenases.
J.Biol.Chem., 296, 2020
|
|
5RLJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1407673036 | | Descriptor: | (2S)-2-phenylpropane-1-sulfonamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM2
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1741964527 | | Descriptor: | 1-(diphenylmethyl)azetidin-3-ol, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMH
 
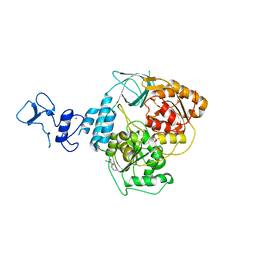 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1101755952 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RL8
 
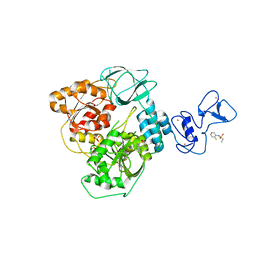 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z53825177 | | Descriptor: | Helicase, N-(2-fluorophenyl)ethanesulfonamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLQ
 
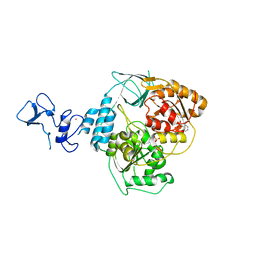 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z285782452 | | Descriptor: | Helicase, N-methyl-2-(methylsulfonyl)aniline, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM9
 
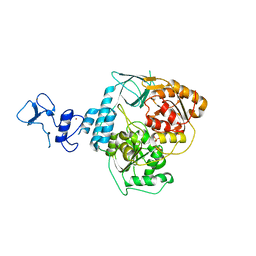 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z2856434942 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z45617795 | | Descriptor: | Helicase, N-(2-phenylethyl)methanesulfonamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLT
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z53116498 | | Descriptor: | 3-(2-methyl-1H-benzimidazol-1-yl)propanamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z2856434920 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLK
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1509882419 | | Descriptor: | 1-(propan-2-yl)-1H-imidazole-4-sulfonamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z216450634 | | Descriptor: | Helicase, N-cycloheptyl-N-methylmethanesulfonamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
