5J0Q
 
 | |
5J0T
 
 | |
5J2I
 
 | |
5J2H
 
 | |
6GEL
 
 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
5J2B
 
 | |
5J2M
 
 | | HIV-1 reverse transcriptase in complex with DNA and EFdA-triphosphate, a translocation-defective RT inhibitor | | Descriptor: | 2'-deoxy-4'-ethynyl-2-fluoroadenosine 5'-(tetrahydrogen triphosphate), DNA (27-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Salie, Z.L, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.432 Å) | | Cite: | Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J2A
 
 | |
5J2F
 
 | |
5J2P
 
 | | HIV-1 reverse transcriptase in complex with DNA that has incorporated EFdA-MP at the P-(post-translocation) site and a second EFdA-MP at the N-(pre-translocation) site | | Descriptor: | 2'-deoxy-4'-ethynyl-2-fluoroadenosine 5'-(dihydrogen phosphate), DNA (27-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(6FM)P*(6FM))-3'), ... | | Authors: | Salie, Z.L, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J6H
 
 | |
6ULX
 
 | |
1HSH
 
 | |
6VBX
 
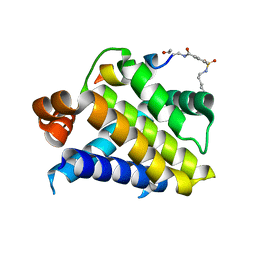 | | Crystal structure of Mcl-1 in complex with 138E12 peptide, Lys-covalent antagonist | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Synthetic peptide | | Authors: | Pellecchia, M, Perry, J.J, Kenjic, N, Assar, Z. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Structural Characterization of Lysine Covalent BH3 Peptides Targeting Mcl-1.
J.Med.Chem., 64, 2021
|
|
1IGI
 
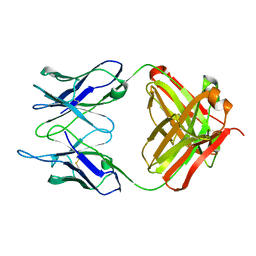 | |
1HSI
 
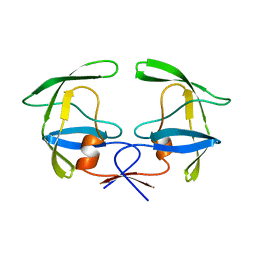 | |
1HSG
 
 | |
1HPC
 
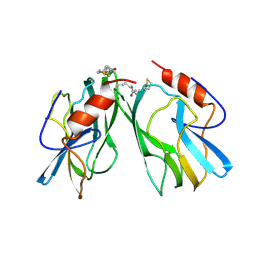 | | REFINED STRUCTURES AT 2 ANGSTROMS AND 2.2 ANGSTROMS OF THE TWO FORMS OF THE H-PROTEIN, A LIPOAMIDE-CONTAINING PROTEIN OF THE GLYCINE DECARBOXYLASE | | Descriptor: | 5-[(3S)-1,2-dithiolan-3-yl]pentanoic acid, H PROTEIN OF THE GLYCINE CLEAVAGE SYSTEM, LIPOIC ACID | | Authors: | Pares, S, Cohen-Addad, C, Sieker, L, Neuburger, M, Douce, R. | | Deposit date: | 1994-02-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures at 2 and 2.2 A resolution of two forms of the H-protein, a lipoamide-containing protein of the glycine decarboxylase complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1IGJ
 
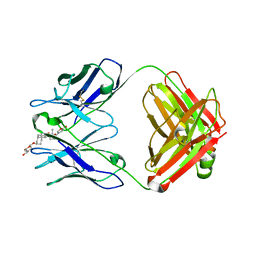 | |
1JXP
 
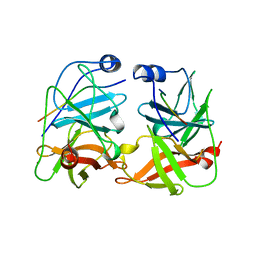 | | BK STRAIN HEPATITIS C VIRUS (HCV) NS3-NS4A | | Descriptor: | NS3 SERINE PROTEASE, NS4A, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-08-21 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
7SSF
 
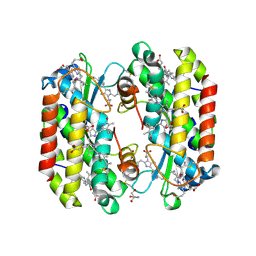 | | Light harvesting phycobiliprotein HaPE560 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DiCys-(15,16)-Dihydrobiliverdin, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii
Commun Biol, 2023
|
|
7SUT
 
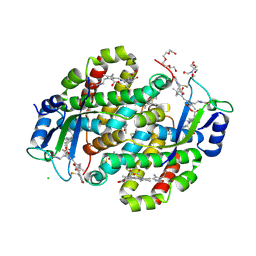 | | Light harvesting phycobiliprotein HaPE645 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | (15,16)-DIHYDROBILIVERDIN (SINGLY LINKED), 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-18 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
1PWW
 
 | |
1PWU
 
 | | Crystal Structure of Anthrax Lethal Factor complexed with (3-(N-hydroxycarboxamido)-2-isobutylpropanoyl-Trp-methylamide), a known small molecule inhibitor of matrix metalloproteases. | | Descriptor: | 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, Lethal factor, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PWV
 
 | |
