4N6Y
 
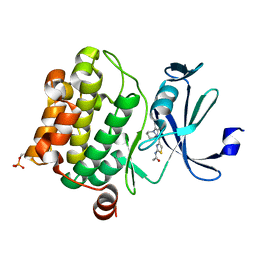 | | Pim1 Complexed with a phenylcarboxamide | | Descriptor: | 2-(acetylamino)-N-[2-(piperidin-1-yl)phenyl]-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
6VVO
 
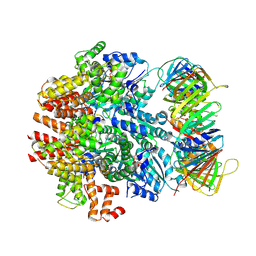 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
8AON
 
 | |
4IPF
 
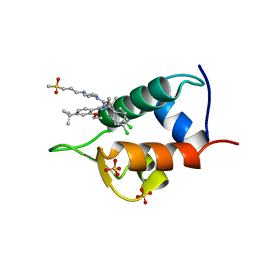 | | The 1.7A crystal structure of humanized Xenopus MDM2 with RO5045337 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(4S,5R)-2-(4-tert-butyl-2-ethoxyphenyl)-4,5-bis(4-chlorophenyl)-4,5-dimethyl-4,5-dihydro-1H-imidazol-1-yl]{4-[3-(methylsulfonyl)propyl]piperazin-1-yl}methanone | | Authors: | Graves, B.J, Lukacs, C, Kammlott, R.U, Crowther, R. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | MDM2 Small-Molecule Antagonist RG7112 Activates p53 Signaling and Regresses Human Tumors in Preclinical Cancer Models.
Cancer Res., 73, 2013
|
|
4IEG
 
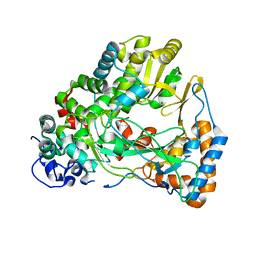 | |
4NW6
 
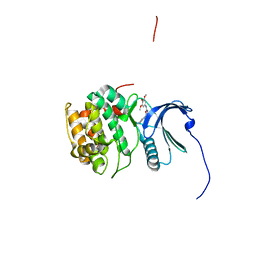 | |
4N1I
 
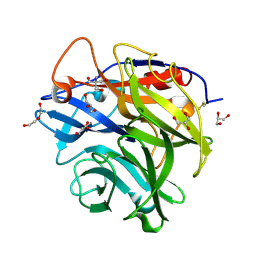 | | Crystal Structure of the alpha-L-arabinofuranosidase UmAbf62A from Ustilago maidys | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-04 | | Release date: | 2014-01-15 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
1LOU
 
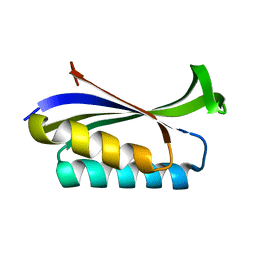 | | RIBOSOMAL PROTEIN S6 | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Otzen, D.E, Kristensen, O, Proctor, M, Oliveberg, M. | | Deposit date: | 1998-11-25 | | Release date: | 1998-11-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural changes in the transition state of protein folding: alternative interpretations of curved chevron plots.
Biochemistry, 38, 1999
|
|
2WWP
 
 | | Crystal structure of the human lipocalin-type prostaglandin D synthase | | Descriptor: | CHLORIDE ION, PROSTAGLANDIN-H2 D-ISOMERASE, THIOCYANATE ION | | Authors: | Roos, A.K, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Dynamic Insights Into Substrate Binding and Catalysis of Human Lipocalin Prostaglandin D Synthase.
J.Lipid Res., 54, 2013
|
|
5SFA
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 6-cyclopropyl-3-[(1-methyl-4-phenylimidazol-2-yl)methoxy]-N-(oxolan-3-yl)pyrazine-2-carboxamide | | Descriptor: | 6-cyclopropyl-3-{[(4S)-1-methyl-4-phenyl-4,5-dihydro-1H-imidazol-2-yl]methoxy}-N-[(3R)-oxolan-3-yl]pyrazine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
3GOY
 
 | | Crystal structure of human poly(adp-ribose) polymerase 14, catalytic fragment in complex with an inhibitor 3-aminobenzamide | | Descriptor: | 3-aminobenzamide, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
4CFK
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH LY294002 | | Descriptor: | 1,2-ETHANEDIOL, 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, BRD4 PROTEIN, ... | | Authors: | Chung, C, Dittmann, A, Drewes, G. | | Deposit date: | 2013-11-18 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Commonly Used Pi3-Kinase Probe Ly294002 is an Inhibitor of Bet Bromodomains.
Acs Chem.Biol., 9, 2014
|
|
4NW5
 
 | |
7Y6D
 
 | |
3H9U
 
 | | S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Siponen, M.I, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kragh Nielsen, T, Kotenyova, T, Kotzsch, A, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei
To be Published
|
|
3TQV
 
 | | Structure of the nicotinate-nucleotide pyrophosphorylase from Francisella tularensis. | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase, PHOSPHATE ION | | Authors: | Rudolph, M, Cheung, J, Franklin, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Rapid countermeasure discovery against Francisella tularensis based on a metabolic network reconstruction.
Plos One, 8, 2013
|
|
4C9X
 
 | | Crystal structure of NUDT1 (MTH1) with S-crizotinib | | Descriptor: | 3-[(1S)-1-(2,6-DICHLORO-3-FLUOROPHENYL)ETHOXY]-5-(1-PIPERIDIN-4-YLPYRAZOL-4-YL)PYRIDIN-2-AMINE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
2ZGX
 
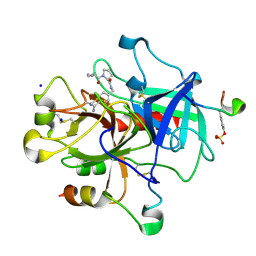 | | Thrombin Inhibition | | Descriptor: | 1-[(2R)-2-aminobutanoyl]-N-(4-carbamimidoylbenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-28 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
2ZDA
 
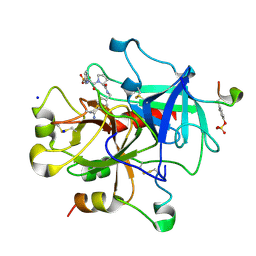 | | Exploring Thrombin S1 pocket | | Descriptor: | D-phenylalanyl-N-{4-[amino(iminio)methyl]benzyl}-L-prolinamide, Hirudin variant-1, PHOSPHATE ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2007-11-21 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
2ZFP
 
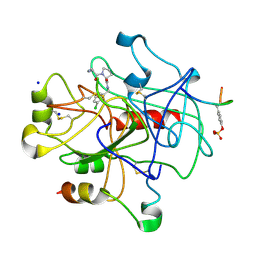 | | Thrombin Inibition | | Descriptor: | 1-[(2R)-2-aminobutanoyl]-N-(3-chlorobenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
2O7A
 
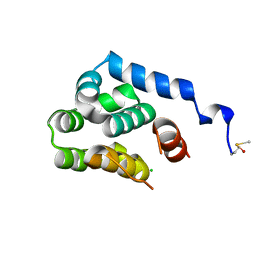 | | T4 lysozyme C-terminal fragment | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme | | Authors: | Echols, N, Kwon, E, Marqusee, S.M, Alber, T. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
3OEQ
 
 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, with full length n-terminus | | Descriptor: | Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
3OER
 
 | | Crystal structure of trimeric frataxin from the yeast saccharomyces cerevisiae, complexed with cobalt | | Descriptor: | COBALT (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
4QSL
 
 | |
3S7E
 
 | | Crystal structure of Ara h 1 | | Descriptor: | Allergen Ara h 1, clone P41B, CHLORIDE ION | | Authors: | Chruszcz, M, Maleki, S.J, Solberg, R, Minor, W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and Immunologic Characterization of Ara h 1, a Major Peanut Allergen.
J.Biol.Chem., 286, 2011
|
|
