3EG0
 
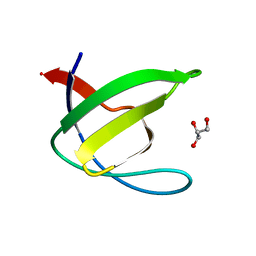 | | Crystal structure of the N114T mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
1YVO
 
 | |
3IGO
 
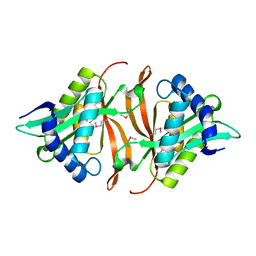
 | | Crystal structure of Cryptosporidium parvum CDPK1, cgd3_920 | | Descriptor: | CALCIUM ION, Calmodulin-domain protein kinase 1, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Finnerty, P, Amani, M, Allali-Hassanali, A, Vedadi, M, Tempel, W, MacKenzie, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7OE2
 
 | | Model of closed pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction | | Descriptor: | Haliangium ochraceum Encapsulated ferritin localisation sequence, Linocin_M18 bacteriocin protein | | Authors: | Marles-Wright, J, Basle, A, Clarke, D.J, Ross, J. | | Deposit date: | 2021-05-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment.
Sci Adv, 8, 2022
|
|
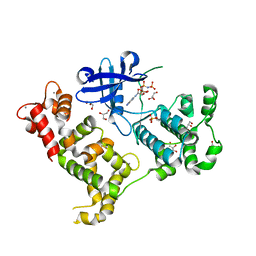
4CKZ
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
8P9R
 
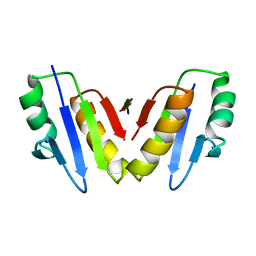 | | Structure of the periplasmic domain of ExbD from E. coli in complex with TonB | | Descriptor: | Biopolymer transport protein ExbD, Protein TonB | | Authors: | Zinke, M, Mechaly, A, Lejeune, M, Haouz, A, Izadi-Pruneyre, N. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ton motor conformational switch and peptidoglycan role in bacterial nutrient uptake.
Nat Commun, 15, 2024
|
|
8Q3U
 
 | | Crystal structure of a fentanyl derivative in complex with human CA VII | | Descriptor: | Carbonic anhydrase 7, GLYCEROL, ZINC ION, ... | | Authors: | Alterio, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2023-08-04 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of a novel series of potent carbonic anhydrase inhibitors with selective affinity for mu Opioid receptor for Safer and long-lasting analgesia.
Eur.J.Med.Chem., 260, 2023
|
|
9ES7
 
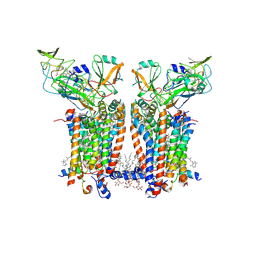 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with water molecules at 1.94 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9ES8
 
 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f with decylplastoquinone bound at plastoquionol reduction site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9ES9
 
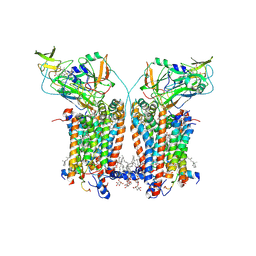 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with inhibitor DBMIB bound at plastoquinol oxidation site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
3ILB
 
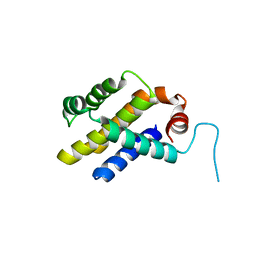 | |
1U4R
 
 | | Crystal Structure of human RANTES mutant 44-AANA-47 | | Descriptor: | SULFATE ION, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
8TBV
 
 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) neoantigen KLSHQLVLL | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
5IS4
 
 | |
3IB0
 
 | | Structural basis of the prevention of NSAID-induced damage of the gastrointestinal tract by C-terminal half (C-lobe) of bovine colostrum protein lactoferrin: Binding and structural studies of C-lobe complex with diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structural basis for the prevention of nonsteroidal antiinflammatory drug-induced gastrointestinal tract damage by the C-lobe of bovine colostrum lactoferrin
Biophys.J., 97, 2009
|
|
10MH
 
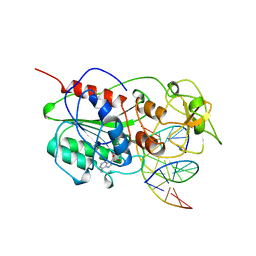 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH ADOHCY AND HEMIMETHYLATED DNA CONTAINING 5,6-DIHYDRO-5-AZACYTOSINE AT THE TARGET | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*5NCP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), ... | | Authors: | Sheikhnejad, G, Brank, A, Christman, J.K, Goddard, A, Alvarez, E, Ford Junior, H, Marquez, V.E, Marasco, C.J, Sufrin, J.R, O'Gara, M, Cheng, X. | | Deposit date: | 1998-08-10 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine.
J.Mol.Biol., 285, 1999
|
|
4LTE
 
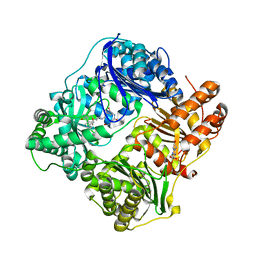 | | Structure of Cysteine-free Human Insulin Degrading Enzyme in Complex with Macrocyclic Inhibitor | | Descriptor: | 2,6-DIAMINO-HEXANOIC ACID AMIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FUMARIC ACID, ... | | Authors: | Foda, Z.H, Seeliger, M.A, Saghatelian, A, Liu, D.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones.
Nature, 511, 2014
|
|
7ODW
 
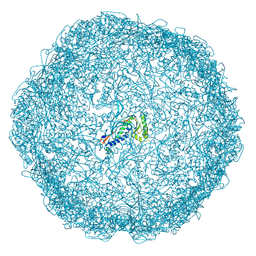 | |
4CZU
 
 | |
4D28
 
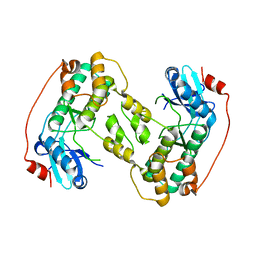 | |
3IAZ
 
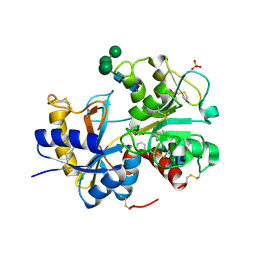 | | Structural basis of the prevention of NSAID-induced damage of the gastrointestinal tract by C-terminal half (C-lobe) of bovine colostrum protein lactoferrin: Binding and structural studies of the C-lobe complex with aspirin | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for the prevention of nonsteroidal antiinflammatory drug-induced gastrointestinal tract damage by the C-lobe of bovine colostrum lactoferrin
Biophys.J., 97, 2009
|
|
5F35
 
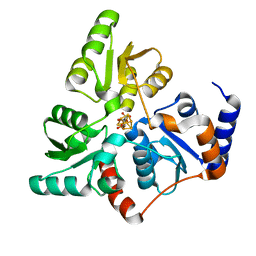 | | Structure of quinolinate synthase in complex with citrate | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
8POC
 
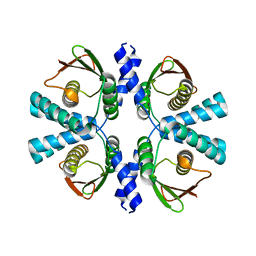 | | Cryo-EM structure of Dickeya dadantii BcsD | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Notopoulou, A, Krasteva, P.V. | | Deposit date: | 2023-07-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures and roles of BcsD and partner scaffold proteins in proteobacterial cellulose secretion.
Curr.Biol., 34, 2024
|
|
5F3D
 
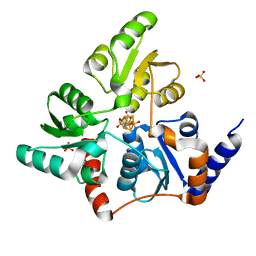 | | Structure of quinolinate synthase in complex with reaction intermediate W | | Descriptor: | 2-IMINO,3-CARBOXY,5-OXO,6-HYDROXY HEXANOIC ACID, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
