7XUH
 
 | | Down-regulated in adenoma in complex with TQR1122 | | Descriptor: | 2-[4,8-dimethyl-2-oxidanylidene-7-[[3-(trifluoromethyl)phenyl]methoxy]chromen-3-yl]ethanoic acid, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | structure of human SLC26A3 in complex with TQR1122
To Be Published
|
|
9IKV
 
 | | human SLC26A7 with iodide | | Descriptor: | Anion exchange transporter, IODIDE ION | | Authors: | Li, X, Yang, X, Lu, X, Lin, B, Zhang, Y, Huang, B, Zhou, Y, Huang, J, Wu, K, Zhou, Q, Chi, X. | | Deposit date: | 2024-06-29 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | human SLC26A7 with iodide
To Be Published
|
|
9IKX
 
 | | human SLC26A7 apo state | | Descriptor: | Anion exchange transporter | | Authors: | Li, X, Yang, X, Lu, X, Lin, B, Zhang, Y, Huang, B, Zhou, Y, Huang, J, Wu, K, Zhou, Q, Chi, X. | | Deposit date: | 2024-06-29 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | human SLC26A7 apo state
To Be Published
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6A69
 
 | | Cryo-EM structure of a P-type ATPase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroplastin, Plasma membrane calcium-transporting ATPase 1 | | Authors: | Gong, D.S, Chi, X.M, Ren, K, Huang, G.X.Y, Zhou, G.W, Yan, N, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-06-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin.
Nat Commun, 9, 2018
|
|
4F1P
 
 | | Crystal Structure of mutant S554D for ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | Deposit date: | 2012-05-07 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
4G1T
 
 | | Crystal structure of interferon-stimulated gene 54 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 2 | | Authors: | Yang, Z, Liang, H, Zhou, Q, Li, Y, Chen, H, Ye, W, Chen, D, Fleming, J, Shu, H, Liu, Y. | | Deposit date: | 2012-07-11 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms.
Cell Res., 22, 2012
|
|
8XL2
 
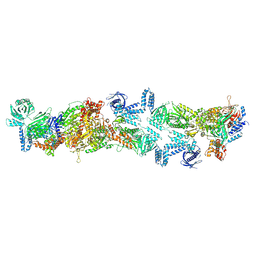 | | Human acetyl-CoA carboxylase 1 filament in complex with acetyl-CoA (ACC1-inact) | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA carboxylase 1 | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XL7
 
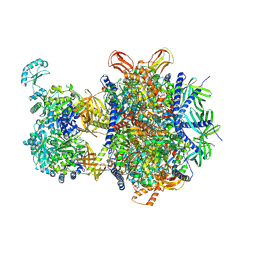 | | Structure of human 3-methylcrotonyl-CoA carboxylase in complex with acetyl-CoA (MCC-ACO) | | Descriptor: | ACETYL COENZYME *A, BIOTIN, Methylcrotonoyl-CoA carboxylase beta chain, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL1
 
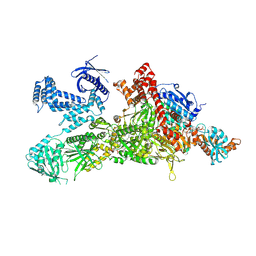 | | Core region of the human acetyl-CoA carboxylase 1 filament in complex with acetyl-CoA (ACC1-inact) | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA carboxylase 1 | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XL8
 
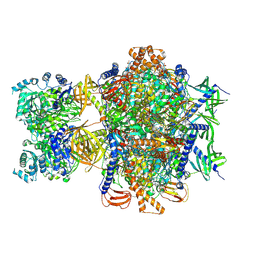 | | Structure of human 3-methylcrotonyl-CoA carboxylase in complex with propionyl-CoA (MCC-PCO) | | Descriptor: | BIOTIN, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL0
 
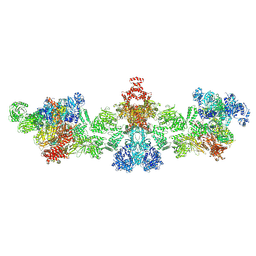 | | Citrate-induced filament of human acetyl-coenzyme A carboxylase 1 (ACC1-citrate) | | Descriptor: | Acetyl-CoA carboxylase 1, BIOTIN | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XKZ
 
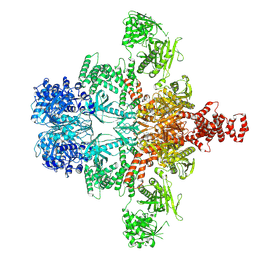 | | Core region of the citrate-induced human acetyl-CoA carboxylase 1 filament (ACC1-citrate) | | Descriptor: | Acetyl-CoA carboxylase 1, BIOTIN | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XL3
 
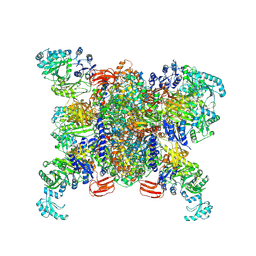 | | Structure of human propionyl-CoA carboxylase at apo-state (PCC-Apo) | | Descriptor: | BIOTIN, Propionyl-CoA carboxylase alpha chain, mitochondrial, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL5
 
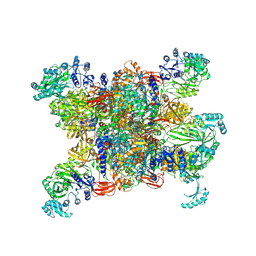 | | Structure of human propionyl-CoA carboxylase in complex with propionyl-CoA (PCC-PCO) | | Descriptor: | BIOTIN, Propionyl-CoA carboxylase alpha chain, mitochondrial, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL6
 
 | | Structure of human 3-methylcrotonyl-CoA carboxylase at apo-state (MCC-Apo) | | Descriptor: | BIOTIN, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL4
 
 | | Structure of human propionyl-CoA carboxylase in complex with acetyl-CoA (PCC-ACO) | | Descriptor: | ACETYL COENZYME *A, BIOTIN, Propionyl-CoA carboxylase alpha chain, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
9J1P
 
 | | Cryo-EM structure of the g1:Ox-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fan, S, Li, J, Zhuang, J, Zhou, Q, Mai, Y, Lin, B, Wang, M.-W, Wu, C. | | Deposit date: | 2024-08-05 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide-Directed Multicyclic Peptides with N-Terminally Extendable alpha-Helices for Recognition and Activation of G Protein-Coupled Receptors.
J.Am.Chem.Soc., 147, 2025
|
|
9JVX
 
 | | Overall structure of human EAAT2 bound with activator (GT949) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 3-[(~{R})-(4-cyclohexylpiperazin-1-yl)-[1-(2-phenylethyl)-1,2,3,4-tetrazol-5-yl]methyl]-6-methoxy-1~{H}-quinolin-2-one, CHOLESTEROL, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Shi, Y, Huang, J, Zhou, Q. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural study of human glutamate transporter EAAT2
Dian Zi Xian Wei Xue Bao, 2025
|
|
8XL9
 
 | | Structure of human pyruvate carboxylase | | Descriptor: | BIOTIN, Pyruvate carboxylase, mitochondrial | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC).
Elife, 2024
|
|
6A70
 
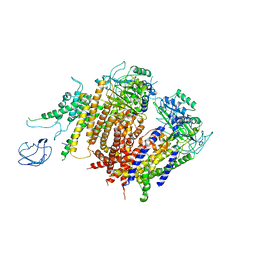 | | Structure of the human PKD1/PKD2 complex | | Descriptor: | Polycystin-1, Polycystin-2 | | Authors: | Su, Q, Hu, F, Ge, X, Lei, J, Yu, S, Wang, T, Zhou, Q, Mei, C, Shi, Y. | | Deposit date: | 2018-06-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the human PKD1-PKD2 complex.
Science, 361, 2018
|
|
8JCB
 
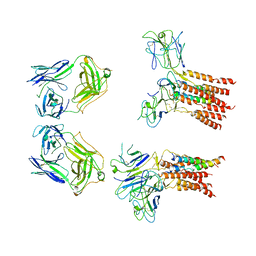 | | Vgamma5 Vdelta1 T cell receptor complex | | Descriptor: | T cell receptor delta variable 1,T cell receptor delta constant, T cell receptor gamma variable 5,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
