4N7R
 
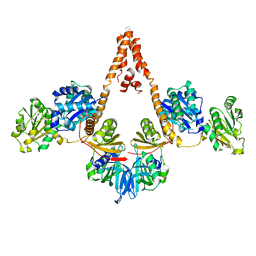 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | Descriptor: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | Authors: | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | Deposit date: | 2013-10-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7OIL
 
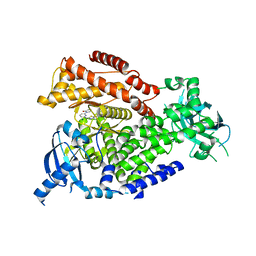 | | mPI3Kd in complex with compound 58 | | Descriptor: | 2-[(1S)-1-cyclopropylethyl]-5-[4-methyl-2-[[6-(2-oxidanylidenepyrrolidin-1-yl)pyridin-2-yl]amino]-1,3-thiazol-5-yl]-7-methylsulfonyl-3H-isoindol-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OI4
 
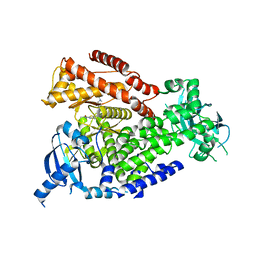 | | mPI3Kd in complex with compound 12 | | Descriptor: | N-[5-[2-[(1S)-1-cyclopropylethyl]-7-[[4-[(dimethylamino)methyl]phenyl]sulfamoyl]-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OIS
 
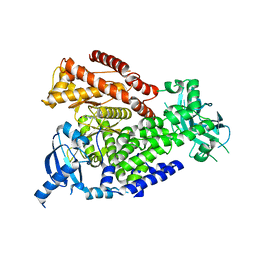 | | mPI3Kd in complex with compound 7 | | Descriptor: | N-[5-[2-[(1S)-1-cyclopropylethyl]-7-[(3-methylsulfonylphenyl)sulfamoyl]-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OIJ
 
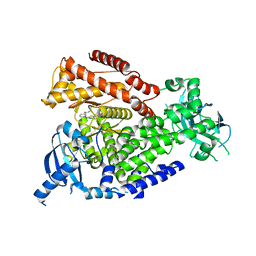 | | mPI3Kd in complex with an inhibitor | | Descriptor: | 6-[[5-[2-[(1S)-1-cyclopropylethyl]-7-(methylsulfamoyl)-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]amino]-N-[3-(dimethylamino)propyl]pyridine-2-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION, ... | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
4QDJ
 
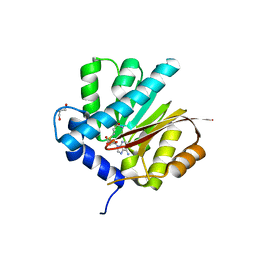 | |
4QDK
 
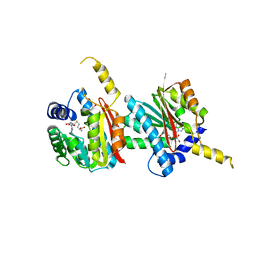 | |
4PS4
 
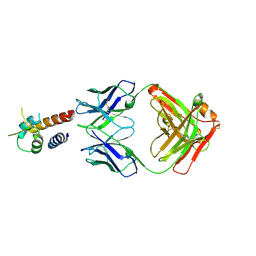 | | Crystal structure of the complex between IL-13 and M1295 FAB | | Descriptor: | Interleukin-13, M1295 HEAVY CHAIN, M1295 LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2014-03-06 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Framework Adaptation of a Mouse Anti-Human Il-13 Antibody.
J.Mol.Biol., 398, 2010
|
|
9J0K
 
 | | An agonist(compound 14e)of Thyroid Hormone Receptor B | | Descriptor: | 2-[[7-[3-(aminomethyl)phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B, Li, Y. | | Deposit date: | 2024-08-02 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Discovery of a Potent, Selective, and Multiple His435 Mutation-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 2025
|
|
9IX5
 
 | | An agonist(compound 15n) of Thyroid Hormone Receptor B | | Descriptor: | 2-[(1-methoxy-7-naphthalen-2-yloxy-4-oxidanyl-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B, Li, Y. | | Deposit date: | 2024-07-26 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a Potent, Selective, and Multiple His435 Mutation-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 2025
|
|
5JYK
 
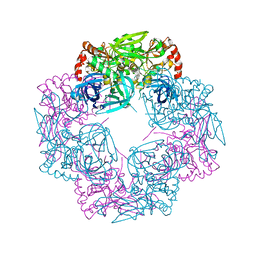 | | Deg9 crystal under 289K | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Zhang, L.X. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
3L5W
 
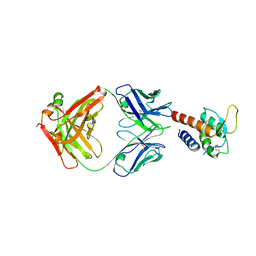 | | Crystal structure of the complex between IL-13 and C836 FAB | | Descriptor: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3L7F
 
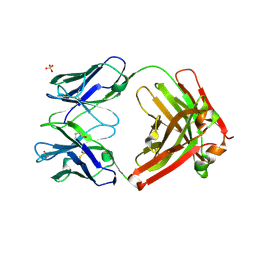 | | Structure of IL-13 antibody H2L6, A humanized variant OF C836 | | Descriptor: | CALCIUM ION, H2L6 HEAVY CHAIN, H2L6 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3L5X
 
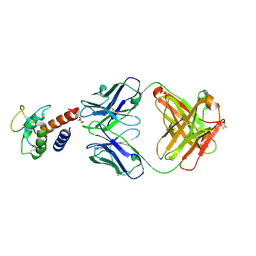 | | Crystal structure of the complex between IL-13 and H2L6 FAB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, H2L6 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
6U84
 
 | |
6U86
 
 | |
6U8A
 
 | |
6U88
 
 | |
5NCY
 
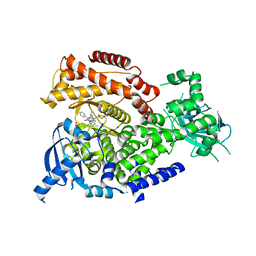 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-(3-methyl-5-oxidanylidene-6-phenyl-[1,3]thiazolo[3,2-a]pyridin-7-yl)ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ... | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
5NCZ
 
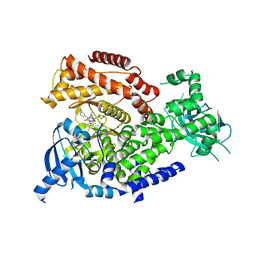 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-[6-[3-[(dimethylamino)methyl]phenyl]-3-methyl-5-oxidanylidene-[1,3]thiazolo[3,2-a]pyridin-7-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
3R22
 
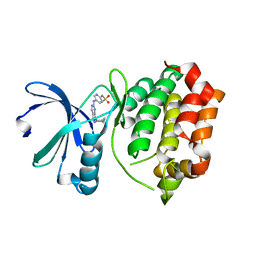 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesana, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R21
 
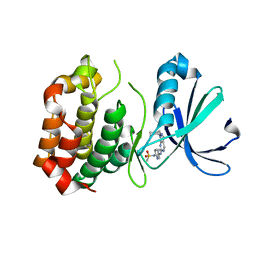 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5XJ6
 
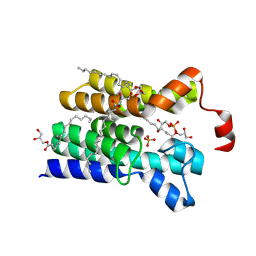 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the glycerol 3-phosphate form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5XJ8
 
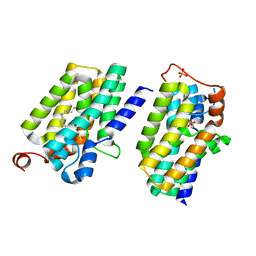 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the lysphosphatidic acid form | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5Y09
 
 | |
