8VKJ
 
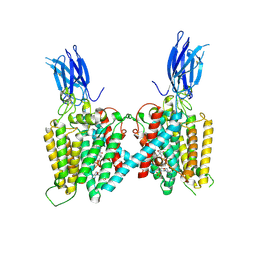 | | Cryo-EM structure of human HGSNAT bound with Acetyl-CoA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VLI
 
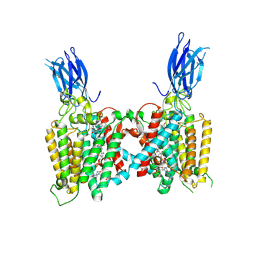 | | Cryo-EM structure of human HGSNAT bound with CoA and product analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methyl-2-oxo-2H-1-benzopyran-7-yl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ... | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VLG
 
 | | Cryo-EM structure of human HGSNAT bound with Acetyl-CoA and substrate analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methyl-2-oxo-2H-1-benzopyran-7-yl 2-amino-2-deoxy-beta-D-glucopyranoside, ... | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VLU
 
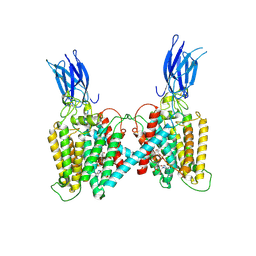 | | Cryo-EM structure of human HGSNAT bound with CoA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COENZYME A, Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VLY
 
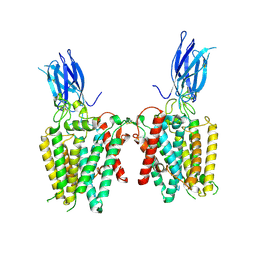 | | Cryo-EM structure of human HGSNAT in transition state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8UJY
 
 | | Crystal structure of human WD repeat-containing protein 5 in complex with 4-(3,5-dimethoxybenzyl)-9-(4-fluoro-2-methylphenyl)-7-((2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl)methyl)-3,4-dihydrobenzo[f][1,4]oxazepin-5(2H)-one (compound 8) | | Descriptor: | (9P)-4-[(3,5-dimethoxyphenyl)methyl]-9-(4-fluoro-2-methylphenyl)-7-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-3,4-dihydro-1,4-benzoxazepin-5(2H)-one, WD repeat-containing protein 5 | | Authors: | Zhao, B, Amporndanai, K, Fesik, S.W. | | Deposit date: | 2023-10-11 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Discovery of Potent, Orally Bioavailable Benzoxazepinone-Based WD Repeat Domain 5 Inhibitors.
J.Med.Chem., 66, 2023
|
|
4HW2
 
 | | Discovery of potent Mcl-1 inhibitors using fragment-based methods and structure-based design | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Zhao, B. | | Deposit date: | 2012-11-07 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design.
J.Med.Chem., 56, 2013
|
|
4HW3
 
 | | Discovery of potent Mcl-1 inhibitors using fragment-based methods and structure-based design | | Descriptor: | 3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-benzothiophene-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2012-11-07 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design.
J.Med.Chem., 56, 2013
|
|
9BRW
 
 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 7 | | Descriptor: | CHLORIDE ION, N-(2-chlorophenyl)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9BRV
 
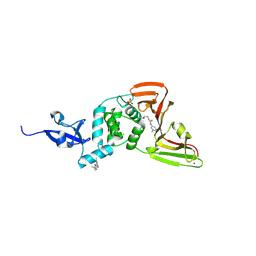 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 5 | | Descriptor: | CHLORIDE ION, N-[2-(dimethylamino)ethyl]-N'-(3-methylphenyl)thiourea, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9BRX
 
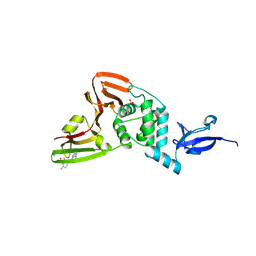 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 10 | | Descriptor: | (4R)-N-(2,4-dimethylphenyl)-7-methyl[1,2,4]triazolo[4,3-a]pyrimidin-5-amine, Papain-like protease nsp3, SULFATE ION, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
4O4S
 
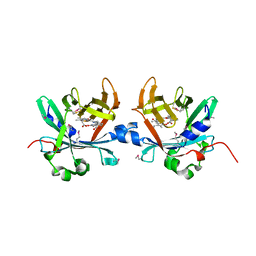 | | Crystal structure of phycobiliprotein lyase CpcT complexed with phycocyanobilin (PCB) | | Descriptor: | PHYCOCYANOBILIN, Phycocyanobilin lyase CpcT | | Authors: | Zhou, W, Ding, W.-L, Zeng, X.-l, Dong, L.-L, Zhao, B, Zhou, M, Scheer, H, Zhao, K.-H, Yang, X. | | Deposit date: | 2013-12-19 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of the Phycobiliprotein Lyase CpcT.
J.Biol.Chem., 289, 2014
|
|
4O4O
 
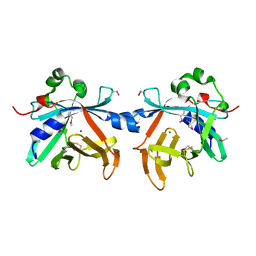 | | Crystal structure of phycobiliprotein lyase CpcT | | Descriptor: | MAGNESIUM ION, Phycocyanobilin lyase CpcT | | Authors: | Zhou, W, Ding, W.-L, Zeng, X.-l, Dong, L.-L, Zhao, B, Zhou, M, Scheer, H, Zhao, K.-H, Yang, X. | | Deposit date: | 2013-12-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Mechanism of the Phycobiliprotein Lyase CpcT.
J.Biol.Chem., 289, 2014
|
|
1NLJ
 
 | |
1NL6
 
 | |
9OMA
 
 | | Cryo-EM structure of PCMTD1-ELOBC-CUL5-RBX2 (CRL5-PCMTD1) | | Descriptor: | Cullin-5, Elongin-B, Elongin-C, ... | | Authors: | Pang, E.Z, Zhao, B, Flowers, C, Oroudjeva, E, Winters, J.B, Pandey, V, Sawaya, M.R, Wohlschlegel, W, Loo, J.A, Rodriguez, J.A, Clarke, S.G. | | Deposit date: | 2025-05-13 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural basis for L-isoaspartyl-containing protein recognition by the PCMTD1 cullin-RING E3 ubiquitin ligase.
Biorxiv, 2025
|
|
9OMF
 
 | | Cryo-EM structure of neddylated PCMTD1-ELOBC-CUL5-RBX2 (N8-CRL5-PCMTD1) | | Descriptor: | Cullin-5, Elongin-B, Elongin-C, ... | | Authors: | Pang, E.Z, Zhao, B, Flowers, C, Oroudjeva, E, Winters, J.B, Pandey, V, Sawaya, M.R, Wohlschlegel, W, Loo, J.A, Rodriguez, J.A, Clarke, S.G. | | Deposit date: | 2025-05-13 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (9.72 Å) | | Cite: | Structural basis for L-isoaspartyl-containing protein recognition by the PCMTD1 cullin-RING E3 ubiquitin ligase.
Biorxiv, 2025
|
|
1HPS
 
 | | RATIONAL DESIGN, SYNTHESIS AND CRYSTALLOGRAPHIC ANALYSIS OF A HYDROXYETHYLENE-BASED HIV-1 PROTEASE INHIBITOR CONTAINING A HETEROCYCLIC P1'-P2' AMIDE BOND ISOSTERE | | Descriptor: | 2-[(1R,3S,4S)-1-BENZYL-4-[N-(BENZYLOXYCARBONYL)-L-VALYL]AMINO-3-PHENYLPENTYL]-4(5)-(2-METHYLPROPIONYL)IMIDAZOLE, HIV-1 PROTEASE | | Authors: | Abdel-Meguid, S, Zhao, B. | | Deposit date: | 1994-05-24 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design, synthesis, and crystallographic analysis of a hydroxyethylene-based HIV-1 protease inhibitor containing a heterocyclic P1'--P2' amide bond isostere.
J.Med.Chem., 37, 1994
|
|
1HOS
 
 | |
7JFM
 
 | |
1BGO
 
 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PEPTIDOMIMETIC INHIBITOR | | Descriptor: | 1-[2-(3-BIPHENYL)-4-METHYLVALERYL)]AMINO-3-(2-PYRIDYLSULFONYL)AMINO-2-PROPANONE, CATHEPSIN K | | Authors: | Desjarlais, R.L, Yamashita, D.S, Oh, H.-J, Bondinell, W.E, Uzinskas, I.N, Erhard, K.F, Allen, A.C, Haltiwanger, R.C, Zhao, B, Smith, W.W, Abdel-Meguid, S.S, D'Alessio, K, Janson, C.A, Mcqueney, M.S, Tomaszek, T.A, Levy, M.A, Veber, D.F. | | Deposit date: | 1998-05-29 | | Release date: | 1999-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of X-Ray Co-Crystal Structures and Molecular Modeling to Design Potent and Selective Non-Peptide Inhibitors of Cathepsin K
J.Am.Chem.Soc., 120, 1998
|
|
7JFL
 
 | |
7RCH
 
 | | Crystal structure of NS1-ED of Vietnam influenza A virus in complex with the p85-beta-iSH2 domain of human PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Kim, I, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Energy landscape reshaped by strain-specific mutations underlies epistasis in NS1 evolution of influenza A virus.
Nat Commun, 13, 2022
|
|
4Z8U
 
 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:-ORF2 WITH ATP | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
6U28
 
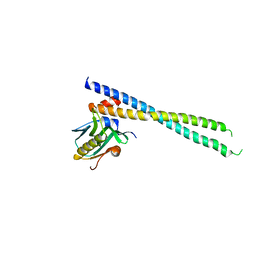 | | Crystal structure of 1918 NS1-ED W187A in complex with the p85-beta-iSH2 domain of human PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Cho, J.H, Zhao, B, Savage, N, Li, P. | | Deposit date: | 2019-08-19 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
