2MED
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEA
 
 | |
2MEB
 
 | | CHANGES IN CONFORMATIONAL STABILITY OF A SERIES OF MUTANT HUMAN LYSOZYMES AT CONSTANT POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEI
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
7V62
 
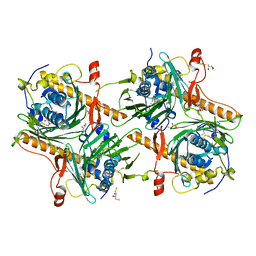 | | Crystal structure of human OSBP ORD in complex with cholesterol | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHOLESTEROL, CITRIC ACID, ... | | Authors: | Kobayashi, J, Kato, R. | | Deposit date: | 2021-08-19 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Ligand Recognition by the Lipid Transfer Domain of Human OSBP Is Important for Enterovirus Replication.
Acs Infect Dis., 8, 2022
|
|
2VLK
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, Van Der Merwe, A, Bell, J, Mcmichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
2VLL
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, Van Der Merwe, A, Bell, J, Mcmichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
2VLR
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, van der Merwe, A, Bell, J, McMichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
2VLJ
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, Van Der Merwe, A, Bell, J, Mcmichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
1L3X
 
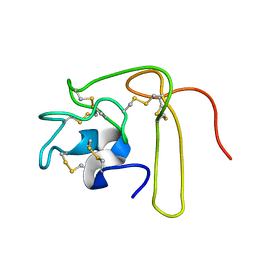 | | Solution Structure of Novel Disintegrin Salmosin | | Descriptor: | platelet aggregation inhibitor disintegrin | | Authors: | Shin, J, Lee, W. | | Deposit date: | 2002-03-01 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel disintegrin, salmosin, from Agkistrondon halys venom
Biochemistry, 42, 2003
|
|
1EJQ
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE | | Descriptor: | SYNDECAN-4 | | Authors: | Shin, J, Oh, E.S, Lee, D, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-04 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE
To be Published
|
|
3W3V
 
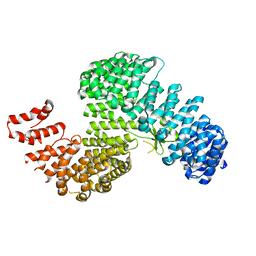 | |
3W3T
 
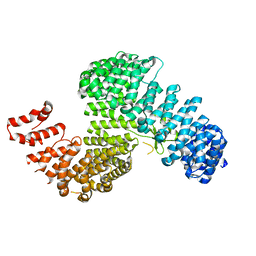 | | Crystal structure of Kap121p | | Descriptor: | Importin subunit beta-3 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3W
 
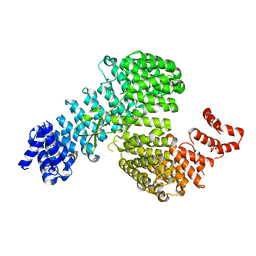 | | Crystal structure of Kap121p bound to Ste12p | | Descriptor: | Importin subunit beta-3, Protein STE12 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3X
 
 | | Crystal structure of Kap121p bound to Pho4p | | Descriptor: | Importin subunit beta-3, Phosphate system positive regulatory protein PHO4 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3Y
 
 | | Crystal structure of Kap121p bound to Nup53p | | Descriptor: | Importin subunit beta-3, Nucleoporin NUP53 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3U
 
 | |
3W3Z
 
 | | Crystal structure of Kap121p bound to RanGTP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin subunit beta-3, ... | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
2LAV
 
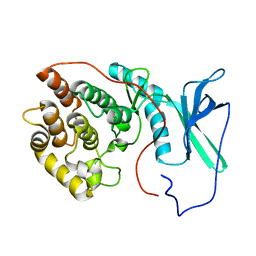 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
2KTY
 
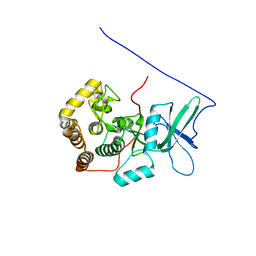 | |
2LJP
 
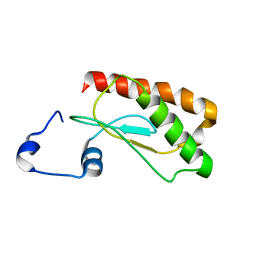 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for E.coli Ribonuclease P protein | | Descriptor: | Ribonuclease P protein component | | Authors: | Shin, J, Kim, K, Ryu, K, Han, K, Lee, Y, Choi, B. | | Deposit date: | 2011-09-21 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Escherichia coli C5 protein
To be Published
|
|
2KUL
 
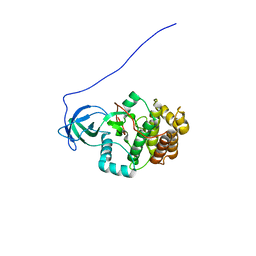 | |
2N5Q
 
 | | Solution structure of cystein-rich peptide jS1 from Jasminum sambac | | Descriptor: | cysteine-rich peptide jS1 | | Authors: | Shin, J, Kumari, G, Serra, A, Nguyen, P.Q.T, Yoon, H, Tam, J.P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification of a cysteine-rich peptide family with unusual disulfide connectivity from Jasminum sambac
To be Published
|
|
3ALM
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant C294A | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
3ALJ
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, reduced form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
