4LCJ
 
 | | CtBP2 in complex with substrate MTOB | | Descriptor: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | Deposit date: | 2013-06-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
5ETX
 
 | | Crystal structure of HCV NS3/4A protease A156T variant in complex with 5172-Linear (MK-5172 linear analogue) | | Descriptor: | CHLORIDE ION, NS3 protease, ZINC ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
8FL5
 
 | |
4U6S
 
 | | CtBP1 in complex with substrate phenylpyruvate | | Descriptor: | 3-PHENYLPYRUVIC ACID, C-terminal-binding protein 1, CALCIUM ION, ... | | Authors: | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
4U6Q
 
 | | CtBP1 bound to inhibitor 2-(hydroxyimino)-3-phenylpropanoic acid | | Descriptor: | (2E)-2-(hydroxyimino)-3-phenylpropanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
9N8P
 
 | | Subtomogram average of dimers of influenza HA trimers | | Descriptor: | Hemagglutinin, Hemagglutinin HA2 chain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose | | Authors: | Huang, Q.J, Song, K, Schiffer, C.A, Somasundaran, M. | | Deposit date: | 2025-02-09 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Virion-associated influenza hemagglutinin clusters upon sialic acid binding visualized by cryoelectron tomography.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5EPY
 
 | | Crystal structure of HCV NS3/4A protease A156T variant in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | Descriptor: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5ESB
 
 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with Vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
5EQR
 
 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
5EQQ
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with 5172-Linear (MK-5172 linear analogue) | | Descriptor: | NS3 protease, SULFATE ION, ZINC ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5EPN
 
 | | Crystal structure of HCV NS3/4A protease in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | Descriptor: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-11 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5EQS
 
 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with Asunaprevir | | Descriptor: | N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, NS3 protease, ZINC ION | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
4WH6
 
 | | Crystal structure of HCV NS3/4A protease variant R155K in complex with Asunaprevir | | Descriptor: | Genome polyprotein, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, SULFATE ION, ... | | Authors: | Soumana, D.I, Ali, A, Schiffer, C.A. | | Deposit date: | 2014-09-20 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Analysis of Asunaprevir Resistance in HCV NS3/4A Protease.
Acs Chem.Biol., 9, 2014
|
|
4WH8
 
 | | Crystal Structure of HCV NS3/4A protease in complex with an Asunaprevir P1-P3 macrocyclic analog. | | Descriptor: | Genome polyprotein, SULFATE ION, ZINC ION, ... | | Authors: | Soumana, D.I, Ali, A, Schiffer, C.A. | | Deposit date: | 2014-09-20 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural Analysis of Asunaprevir Resistance in HCV NS3/4A Protease.
Acs Chem.Biol., 9, 2014
|
|
9MEI
 
 | | Crystal Structure of SARS-CoV-2 Mpro mutant L50F E166V with Pfizer Intravenous Inhibitor PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Zvornicanin, S.N, Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2024-12-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular mechanisms of drug resistance and compensation in SARS-CoV-2 main protease: the interplay between E166 and L50.
Mbio, 16, 2025
|
|
2QI4
 
 | | Crystal structure of protease inhibitor, MIT-2-AD93 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-3-HYDROXYBENZAMIDE, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QI3
 
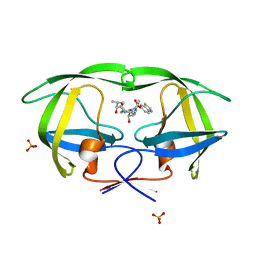 | | Crystal structure of protease inhibitor, MIT-2-AD94 in complex with wild type HIV-1 protease | | Descriptor: | (2S)-N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-2-HYDROXY-3-METHYLBUTANAMIDE, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
5VP9
 
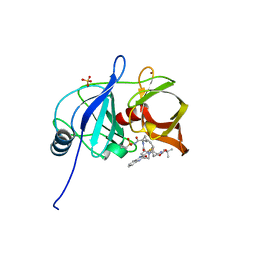 | |
6CVX
 
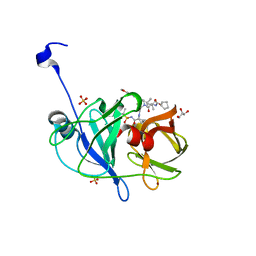 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-50 (MK-5172 linear analogue) | | Descriptor: | GLYCEROL, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-4-{[7-methoxy-3-(propan-2-yl)quinoxalin-2-yl]oxy}-L-prolinamide, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
6CVW
 
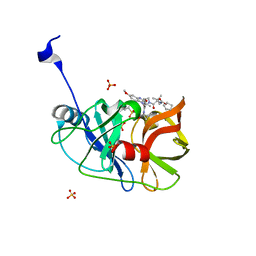 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-52 (MK-5172 linear analogue) | | Descriptor: | N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-4-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-L-prolinamide, NS3 protease, SULFATE ION, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
6CVY
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-21 (MK-5172 linear analogue) | | Descriptor: | 3-methyl-N-[(pentyloxy)carbonyl]-L-valyl-(4R)-4-[(3-chloro-7-methoxyquinoxalin-2-yl)oxy]-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-L-prolinamide, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
5VOJ
 
 | |
8W3C
 
 | |
8W3M
 
 | | Crystal Structure of Enterovirus 68 3C Protease with AG7404 at 1.97 Angstroms | | Descriptor: | Peptidase C3, SODIUM ION, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Authors: | Azzolino, V.N, Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2024-02-22 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Analysis of Inhibitor Binding to Enterovirus-D68 3C Protease.
Viruses, 17, 2025
|
|
8W3T
 
 | |
