1OU9
 
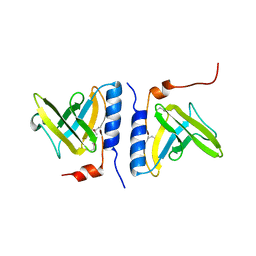 | | Structure of SspB, a AAA+ protease delivery protein | | Descriptor: | CALCIUM ION, Stringent starvation protein B homolog | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
1OUL
 
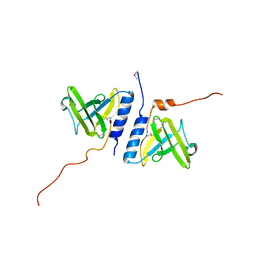 | | Structure of the AAA+ protease delivery protein SspB | | Descriptor: | Stringent starvation protein B homolog | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
1BDT
 
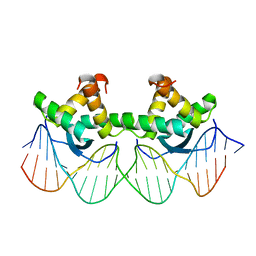 | | WILD TYPE GENE-REGULATING PROTEIN ARC/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*TP*AP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*TP*AP*TP*CP*AP*T)-3'), PROTEIN (GENE-REGULATING PROTEIN ARC) | | Authors: | Schilbach, J.F, Karzai, A.W, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1998-05-11 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Origins of DNA-binding specificity: role of protein contacts with the DNA backbone.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1Q5V
 
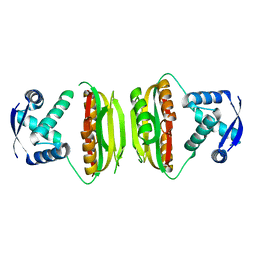 | | Apo-NikR | | Descriptor: | Nickel responsive regulator | | Authors: | Schreiter, E.R, Sintchak, M.D, Guo, Y, Chivers, P.T, Sauer, R.T, Drennan, C.L. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nickel-Responsive Transcription Factor NikR
Nat.Struct.Biol., 10, 2003
|
|
1Q5Y
 
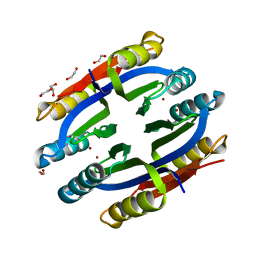 | | Nickel-Bound C-terminal Regulatory Domain of NikR | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Nickel responsive regulator | | Authors: | Schreiter, E.R, Sintchak, M.D, Guo, Y, Chivers, P.T, Sauer, R.T, Drennan, C.L. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Nickel-Responsive Transcription Factor NikR
Nat.Struct.Biol., 10, 2003
|
|
1QTG
 
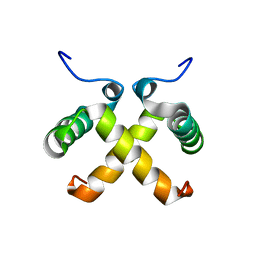 | | AVERAGED NMR MODEL OF SWITCH ARC, A DOUBLE MUTANT OF ARC REPRESSOR | | Descriptor: | Transcriptional repressor arc | | Authors: | Cordes, M.H.J, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | Deposit date: | 1999-06-27 | | Release date: | 1999-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Evolution of a protein fold in vitro.
Science, 284, 1999
|
|
1PAR
 
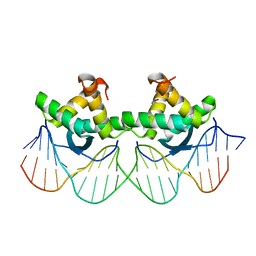 | | DNA RECOGNITION BY BETA-SHEETS IN THE ARC REPRESSOR-OPERATOR CRYSTAL STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*T P*AP*CP*TP*AP*T)- 3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*T P*AP*TP*CP*AP*T)- 3'), PROTEIN (ARC REPRESSOR) | | Authors: | Raumann, B.E, Rould, M.A, Pabo, C.O, Sauer, R.T. | | Deposit date: | 1994-03-22 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA recognition by beta-sheets in the Arc repressor-operator crystal structure.
Nature, 367, 1994
|
|
1MYK
 
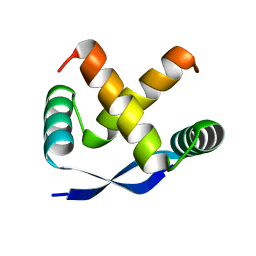 | | CRYSTAL STRUCTURE, FOLDING, AND OPERATOR BINDING OF THE HYPERSTABLE ARC REPRESSOR MUTANT PL8 | | Descriptor: | ARC REPRESSOR | | Authors: | Schildbach, J.F, Milla, M.E, Jeffrey, P.D, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1994-10-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure, folding, and operator binding of the hyperstable Arc repressor mutant PL8.
Biochemistry, 34, 1995
|
|
1MNT
 
 | | SOLUTION STRUCTURE OF DIMERIC MNT REPRESSOR (1-76) | | Descriptor: | MNT REPRESSOR | | Authors: | Burgering, M.J.M, Boelens, R, Gilbert, D.E, Breg, J.N, Knight, K.L, Sauer, R.T, Kaptein, R. | | Deposit date: | 1994-06-28 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dimeric Mnt repressor (1-76).
Biochemistry, 33, 1994
|
|
1MYL
 
 | |
1OU8
 
 | | structure of an AAA+ protease delivery protein in complex with a peptide degradation tag | | Descriptor: | MAGNESIUM ION, Stringent starvation protein B homolog, synthetic ssrA peptide | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
1LLI
 
 | | THE CRYSTAL STRUCTURE OF A MUTANT PROTEIN WITH ALTERED BUT IMPROVED HYDROPHOBIC CORE PACKING | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Lim, W.A, Hodel, A, Sauer, R.T, Richards, F.M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a mutant protein with altered but improved hydrophobic core packing.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1NLA
 
 | | Solution Structure of Switch Arc, a Mutant with 3(10) Helices Replacing a Wild-Type Beta-Ribbon | | Descriptor: | Transcriptional repressor arc | | Authors: | Cordes, M.H, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Switch Arc, a mutant with 3(10) helices replacing a wild-type beta-ribbon
J.Mol.Biol., 326, 2003
|
|
2TBV
 
 | |
1VCW
 
 | | Crystal structure of DegS after backsoaking the activating peptide | | Descriptor: | Protease degS | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease.
Cell(Cambridge,Mass.), 117, 2004
|
|
3ORC
 
 | |
3KZ0
 
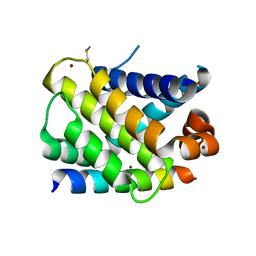 | | MCL-1 complex with MCL-1-specific selected peptide | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 specific peptide MB7, SULFATE ION, ... | | Authors: | Dutta, S, Fire, E, Grant, R.A, Sauer, R.T, Keating, A.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL.
J.Mol.Biol., 398, 2010
|
|
2ORC
 
 | |
1DU0
 
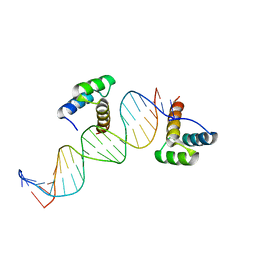 | | ENGRAILED HOMEODOMAIN Q50A VARIANT DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*CP*CP*TP*AP*A)-3'), ENGRAILED HOMEODOMAIN | | Authors: | Grant, R.A, Rould, M.A, Klemm, J.D, Pabo, C.O. | | Deposit date: | 2000-01-13 | | Release date: | 2000-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the role of glutamine 50 in the homeodomain-DNA interface: crystal structure of engrailed (Gln50 --> ala) complex at 2.0 A.
Biochemistry, 39, 2000
|
|
1SOT
 
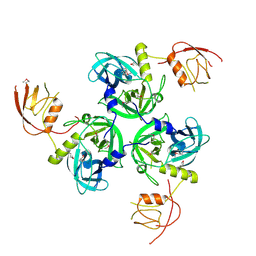 | | Crystal Structure of the DegS stress sensor | | Descriptor: | Protease degS | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1SOZ
 
 | | Crystal Structure of DegS protease in complex with an activating peptide | | Descriptor: | Protease degS, activating peptide | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1LMB
 
 | | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Beamer, L.J, Pabo, C.O. | | Deposit date: | 1991-11-05 | | Release date: | 1991-11-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A crystal structure of the lambda repressor-operator complex.
J.Mol.Biol., 227, 1992
|
|
1LRP
 
 | |
1ORC
 
 | |
