2V0S
 
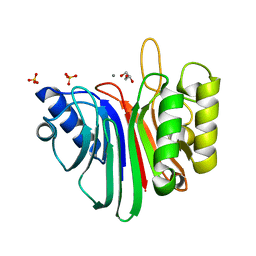 | | crystal structure of a hairpin exchange variant (LR1) of the targeting LINE-1 retrotransposon endonuclease | | Descriptor: | GLYCEROL, LR1, MANGANESE (II) ION, ... | | Authors: | Repanas, K, Zingler, N, Layer, L.E, Schumann, G.G, Perrakis, A, Weichenrieder, O. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Determinants for DNA Target Structure Selectivity of the Human Line-1 Retrotransposon Endonuclease.
Nucleic Acids Res., 35, 2007
|
|
2V0R
 
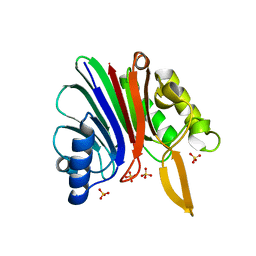 | | crystal structure of a hairpin exchange variant (LTx) of the targeting LINE-1 retrotransposon endonuclease | | Descriptor: | LTX, SULFATE ION | | Authors: | Repanas, K, Zingler, N, Layer, L.E, Schumann, G.G, Perrakis, A, Weichenrieder, O. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determinants for DNA Target Structure Selectivity of the Human Line-1 Retrotransposon Endonuclease
Nucleic Acids Res., 35, 2007
|
|
2X4S
 
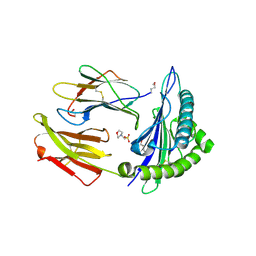 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a peptide representing the epitope of the H5N1 (Avian Flu) Nucleoprotein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals
J.Am.Chem.Soc., 131, 2009
|
|
6QC0
 
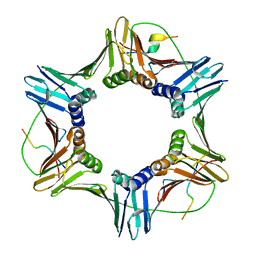 | | PCNA complex with Cdt2 C-terminal PIP-box peptide | | Descriptor: | Denticleless protein homolog, Proliferating cell nuclear antigen | | Authors: | Perrakis, A.P, von Castelmur, E. | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Direct binding of Cdt2 to PCNA is important for targeting the CRL4Cdt2E3 ligase activity to Cdt1.
Life Sci Alliance, 1, 2018
|
|
7OC9
 
 | |
8R88
 
 | |
7M6B
 
 | | The Crystal Structure of Mcbe1 | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2021-03-25 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
8PH4
 
 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
1K9T
 
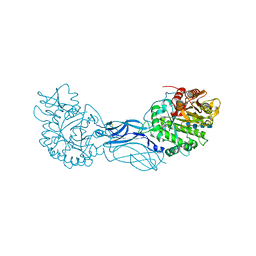 | | Chitinase a complexed with tetra-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Prag, G, Tucker, P.A, Oppenheim, A.B. | | Deposit date: | 2001-10-30 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Structures of Chitinase A Mutant with Oligonag Provide Insight Into the Enzymatic Mechanism
To be Published
|
|
1C7S
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT D539A COMPLEXED WITH DI-N-ACETYL-BETA-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-14 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1SEN
 
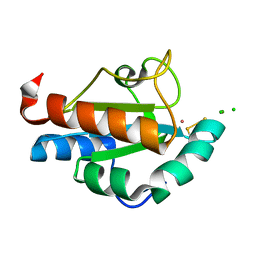 | | Endoplasmic reticulum protein Rp19 O95881 | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, Thioredoxin-like protein p19 | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Lee, D, Dailey, T.A, Mayer, M.R, Rose, J.P, Richardson, D.C, Richardson, J.S, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-07-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Endoplasmic reticulum protein Rp19
To be Published
|
|
8TLY
 
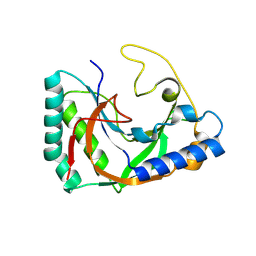 | |
8TUK
 
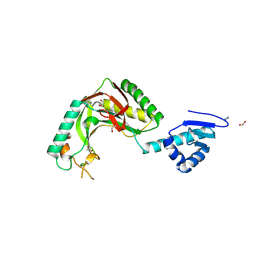 | | Alvinella ASCC1 KH and Phosphodiesterase/Ligase Domain | | Descriptor: | 1,2-ETHANEDIOL, Activating signal cointegrator 1 complex subunit 1, IMIDAZOLE | | Authors: | Tsutakawa, S.E, Tainer, J.A, Arvai, A.S, Chinnam, N.B. | | Deposit date: | 2023-08-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | ASCC1 structures and bioinformatics reveal a novel helix-clasp-helix RNA-binding motif linked to a two-histidine phosphodiesterase.
J.Biol.Chem., 300, 2024
|
|
1DEK
 
 | |
1DEL
 
 | |
8DAF
 
 | | Human SF-1 LBD bound to synthetic agonist 6N-10CA and bacterial phospholipid | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | D'Agostino, E.H, Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparison of activity, structure, and dynamics of SF-1 and LRH-1 complexed with small molecule modulators.
J.Biol.Chem., 299, 2023
|
|
1LTM
 
 | | ACCELERATED X-RAY STRUCTURE ELUCIDATION OF A 36 KDA MURAMIDASE/TRANSGLYCOSYLASE USING WARP | | Descriptor: | 1,2-ETHANEDIOL, 36 KDA SOLUBLE LYTIC TRANSGLYCOSYLASE, BICINE, ... | | Authors: | Van Asselt, E.J, Dijkstra, B.W. | | Deposit date: | 1997-09-26 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accelerated X-ray structure elucidation of a 36 kDa muramidase/transglycosylase using wARP.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1VAM
 
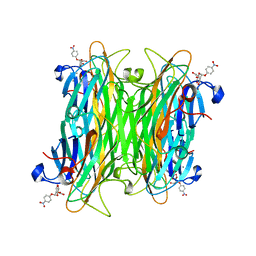 | | CONCANAVALIN A COMPLEX WITH 4'-NITROPHENYL-ALPHA-D-MANNOPYRANOSIDE | | Descriptor: | 4-nitrophenyl alpha-D-mannopyranoside, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Kanellopoulos, P.N, Tucker, P.A, Hamodrakas, S.J. | | Deposit date: | 1996-01-08 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of the complexes of concanavalin A with 4'-nitrophenyl-alpha-D-mannopyranoside and 4'-nitrophenyl-alpha-D-glucopyranoside.
J.Struct.Biol., 116, 1996
|
|
1VAL
 
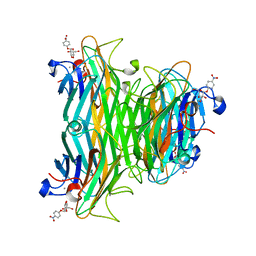 | | CONCANAVALIN A COMPLEX WITH 4'-NITROPHENYL-ALPHA-D-GLUCOPYRANOSIDE | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Kanellopoulos, P.N, Tucker, P.A, Hamodrakas, S.J. | | Deposit date: | 1996-01-08 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the complexes of concanavalin A with 4'-nitrophenyl-alpha-D-mannopyranoside and 4'-nitrophenyl-alpha-D-glucopyranoside.
J.Struct.Biol., 116, 1996
|
|
1MQS
 
 | |
1TRY
 
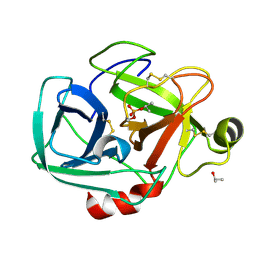 | | STRUCTURE OF INHIBITED TRYPSIN FROM FUSARIUM OXYSPORUM AT 1.55 ANGSTROMS | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHORYLISOPROPANE, TRYPSIN | | Authors: | Rypniewski, W.R, Dambmann, C, Von Der Osten, C, Dauter, M, Wilson, K.S. | | Deposit date: | 1994-03-07 | | Release date: | 1996-01-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of inhibited trypsin from Fusarium oxysporum at 1.55 A.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2IVG
 
 | | SITE DIRECTED MUTAGENESIS OF KEY RESIDUES INVOLVED IN THE CATALYTIC MECHANISM OF CYANASE | | Descriptor: | AZIDE ION, CHLORIDE ION, CYANATE LYASE, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, P.M. | | Deposit date: | 2006-06-13 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
2IU7
 
 | |
2IVB
 
 | | SITE DIRECTED MUTAGENESIS OF KEY RESIDUES INVOLVED IN THE CATALYTIC MECHANISM OF CYANASE | | Descriptor: | AZIDE ION, CHLORIDE ION, CYANATE HYDRATASE, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, P.M. | | Deposit date: | 2006-06-09 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
