1G81
 
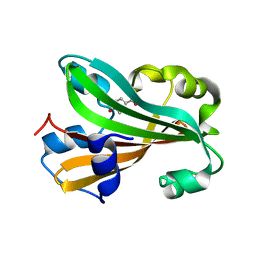 | | CHORISMATE LYASE WITH BOUND PRODUCT, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Vilker, V.L, Howard, A. | | Deposit date: | 2000-11-15 | | Release date: | 2001-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
1V9U
 
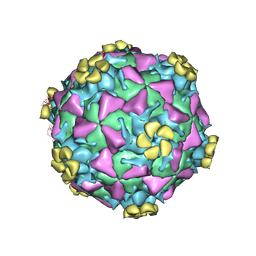 | | Human Rhinovirus 2 bound to a fragment of its cellular receptor protein | | Descriptor: | CALCIUM ION, Coat protein VP1, Coat protein VP2, ... | | Authors: | Verdaguer, N, Fita, I, Reithmayer, M, Moser, R, Blaas, D. | | Deposit date: | 2004-02-03 | | Release date: | 2004-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
5T4M
 
 | |
5TPK
 
 | |
5T4N
 
 | |
5TFL
 
 | | Crystal Structure of Mouse Cadherin-23 EC7+8 | | Descriptor: | CALCIUM ION, Cadherin-23, SODIUM ION | | Authors: | Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2016-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5TFM
 
 | | Crystal Structure of Human Cadherin-23 EC6-8 | | Descriptor: | CALCIUM ION, Cadherin-23, SODIUM ION | | Authors: | Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2016-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5UN2
 
 | |
5ULU
 
 | |
5UZ8
 
 | | Crystal Structure of Mouse Cadherin-23 EC22-24 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin-23, ... | | Authors: | Patel, A, Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5ULY
 
 | |
5TFK
 
 | |
5VH2
 
 | |
3GR8
 
 | | Structure of OYE from Geobacillus kaustophilus, orthorhombic crystal form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase, ... | | Authors: | Uhl, M.K, Gruber, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Old Yellow Enzyme-Catalyzed Dehydrogenation of Saturated Ketones
ADV.SYNTH.CATAL., 353, 2011
|
|
2DWU
 
 | | Crystal Structure of Glutamate Racemase Isoform RacE1 from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, GLYCEROL, Glutamate racemase, ... | | Authors: | Mehboob, S, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2006-08-17 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of Two Glutamate Racemase Isozymes from Bacillus anthracis and Implications for Inhibitor Design
J.Mol.Biol., 371, 2007
|
|
3GR7
 
 | |
6MEQ
 
 | | PcdhgB3 EC1-4 in 50 mM HEPES | | Descriptor: | CALCIUM ION, Protocadherin gamma-B3 | | Authors: | Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MER
 
 | | PcdhgB3 EC1-4 in 50 mM HEPES | | Descriptor: | CALCIUM ION, Protocadherin gamma-B3 | | Authors: | Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6YYK
 
 | | Crystal Structure of 1,5-dimethylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1,5-dimethyl-3~{H}-indol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYF
 
 | | Crystal Structure of 5-chloroindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-chloranyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYG
 
 | | Crystal Structure of 5-(trifluoromethoxy)indoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-(trifluoromethyloxy)-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6TSE
 
 | | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1-methylindole-2,3-dione, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
6TVN
 
 | | Crystal Structure of 5-bromoindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase | | Descriptor: | 5-bromanyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
6TUH
 
 | | The PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 4,5,6,7-tetrahydro-1-benzofuran-3-carboxylic acid, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
6TT2
 
 | |
