6FTJ
 
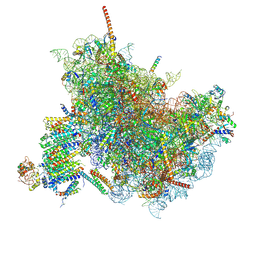 | | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Non-programmed 80S Ribosome | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Braunger, K, Becker, T, Beckmann, R. | | Deposit date: | 2018-02-22 | | Release date: | 2018-03-21 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum.
Science, 360, 2018
|
|
5C37
 
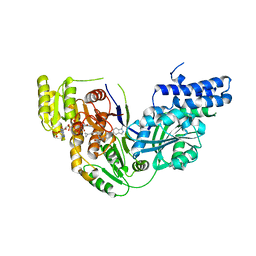 | | Structure of the beta-ketoacyl reductase domain of human fatty acid synthase bound to a spiro-imidazolone inhibitor | | Descriptor: | 6-{[(3R)-1-(cyclopropylcarbonyl)pyrrolidin-3-yl]methyl}-5-[4-(1-methyl-1H-indazol-5-yl)phenyl]-4,6-diazaspiro[2.4]hept-4-en-7-one, CHLORIDE ION, Fatty acid synthase, ... | | Authors: | Schubert, C, Milligan, C.M, Vo, K, Grasberger, B. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of a series of bioavailable fatty acid synthase (FASN) KR domain inhibitors for cancer therapy.
Bioorg.Med.Chem.Lett., 28, 2018
|
|
1B53
 
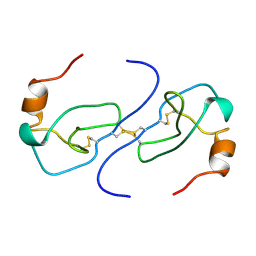 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
1B50
 
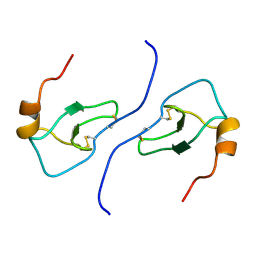 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, 10 STRUCTURES | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
6FTG
 
 | |
3HDN
 
 | |
3HDM
 
 | |
3II9
 
 | | Crystal structure of glutaryl-coa dehydrogenase from Burkholderia pseudomallei at 1.73 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glutaryl-CoA dehydrogenase, ... | | Authors: | Ismagilov, R.F, Li, L, Du, W.B, Staker, B, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-07-31 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | User-loaded SlipChip for equipment-free multiplexed nanoliter-scale experiments.
J.Am.Chem.Soc., 132, 2010
|
|
7UXE
 
 | | Pseudomonas phage E217 small terminase (TerS) | | Descriptor: | Small terminase | | Authors: | Lokareddy, R.K, Hou, C.-F.D, Doll, S.G, Li, F, Gillilan, R, Forti, F, Briani, F, Cingolani, G. | | Deposit date: | 2022-05-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Terminase Subunits from the Pseudomonas-Phage E217.
J.Mol.Biol., 434, 2022
|
|
8UR5
 
 | |
8UR7
 
 | |
6OCQ
 
 | | Crystal structure of RIP1 kinase in complex with a pyrrolidine | | Descriptor: | 1-[(2S)-2-(3-fluorophenyl)pyrrolidin-1-yl]-2,2-dimethylpropan-1-one, Receptor-interacting serine/threonine-protein kinase 1, SULFATE ION | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
6OYW
 
 | | ASK1 kinase domain in complex with Compound 11 | | Descriptor: | (19S)-19-methyl-16,17,18,19-tetrahydro-8,4-(azeno)[1,2,4]triazolo[4,3-f][1,6,13]benzoxadiazacyclohexadecin-10(9H)-one, MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Marcotte, D.J. | | Deposit date: | 2019-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Rational Design and Optimization of a Novel Class of Macrocyclic Apoptosis Signal-Regulating Kinase 1 Inhibitors.
J.Med.Chem., 62, 2019
|
|
6P5C
 
 | | Bacillus Fragment DNA polymerase mutant I716M | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*TP*GP*AP*TP*CP*G)-3'), DNA polymerase I, ... | | Authors: | Wu, E.Y. | | Deposit date: | 2019-05-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The importance of Ile716 toward the mutagenicity of 8-Oxo-2'-deoxyguanosine with Bacillus fragment DNA polymerase.
DNA Repair (Amst.), 89, 2020
|
|
6OYT
 
 | | ASK1 kinase domain in complex with GS-4997 | | Descriptor: | 5-(4-cyclopropyl-1H-imidazol-1-yl)-2-fluoro-4-methyl-N-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}benzamide, ACETATE ION, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Marcotte, D.J. | | Deposit date: | 2019-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.824 Å) | | Cite: | Rational Design and Optimization of a Novel Class of Macrocyclic Apoptosis Signal-Regulating Kinase 1 Inhibitors.
J.Med.Chem., 62, 2019
|
|
6VRE
 
 | | Structure of ASK1 bound to BIO-1772961 | | Descriptor: | 3-methoxy-N-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-1-(pyrazin-2-yl)-1H-pyrazole-4-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Chodaparambil, J.V, Marcotte, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of CNS-Penetrant Apoptosis Signal-Regulating Kinase 1 (ASK1) Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6Z8L
 
 | | Alpha-Amylase in complex with probe fragments | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.40000367 Å) | | Cite: | Enhancing glycan stability via site-selective fluorination: modulating substrate orientation by molecular design.
Chem Sci, 12, 2020
|
|
4NEU
 
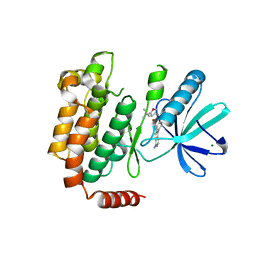 | | X-ray structure of Receptor Interacting Protein 1 (RIP1)kinase domain with a 1-aminoisoquinoline inhibitor | | Descriptor: | 1-[4-(1-aminoisoquinolin-5-yl)phenyl]-3-(5-tert-butyl-1,2-oxazol-3-yl)urea, MAGNESIUM ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Nolte, R.T, Ward, P, kahler, K.M, Campobasso, N. | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of Small Molecule RIP1 Kinase Inhibitors for the Treatment of Pathologies Associated with Necroptosis.
ACS Med Chem Lett, 4, 2013
|
|
5AMV
 
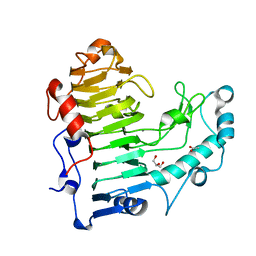 | | Structural insights into the loss of catalytic competence in pectate lyase at low pH | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Teixeira, S.C.M, Ali, S, Sondergaard, C, Pickersgill, R. | | Deposit date: | 2015-09-02 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Insights Into the Loss of Catalytic Competence in Pectate Lyase Activity at Low Ph.
FEBS Lett., 589, 2015
|
|
3HZR
 
 | |
8A1S
 
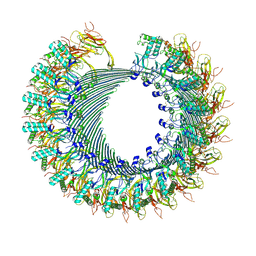 | | Structure of murine perforin-2 (Mpeg1) pore in twisted form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
8A1D
 
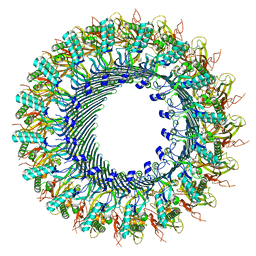 | | Structure of murine perforin-2 (Mpeg1) pore in ring form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
5LZL
 
 | | Pyrobaculum calidifontis 5-aminolaevulinic acid dehydratase | | Descriptor: | Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Azim, N, Erskine, P.T, Guo, J, Cooper, J.B. | | Deposit date: | 2016-09-30 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5MHB
 
 | | Product-Complex of E.coli 5-Amino Laevulinic Acid Dehydratase | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, Delta-aminolevulinic acid dehydratase, GLYCEROL, ... | | Authors: | Norton, E, Erskine, P.T, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2016-11-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3HV0
 
 | |
